8RUD
 
 | | Crystal structure of Rhizobium etli L-asparaginase ReAV K138A mutant | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Pokrywka, K, Grzechowiak, M, Sliwiak, J, Worsztynowicz, P, Loch, J.I, Ruszkowski, M, Gilski, M, Jaskolski, M. | | Deposit date: | 2024-01-30 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Probing the active site of Class 3 L-asparaginase by mutagenesis. I. Tinkering with the zinc coordination site of ReAV.
Front Chem, 12, 2024
|
|
8RUG
 
 | | Crystal structure of Rhizobium etli L-asparaginase ReAV C189A mutant | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Pokrywka, K, Grzechowiak, M, Sliwiak, J, Worsztynowicz, P, Loch, J.I, Ruszkowski, M, Gilski, M, Jaskolski, M. | | Deposit date: | 2024-01-30 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Probing the active site of Class 3 L-asparaginase by mutagenesis. I. Tinkering with the zinc coordination site of ReAV.
Front Chem, 12, 2024
|
|
8RUA
 
 | | Crystal structure of Rhizobium etli L-asparaginase ReAV C135A mutant | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Pokrywka, K, Grzechowiak, M, Sliwiak, J, Worsztynowicz, P, Loch, J.I, Ruszkowski, M, Gilski, M, Jaskolski, M. | | Deposit date: | 2024-01-30 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Probing the active site of Class 3 L-asparaginase by mutagenesis. I. Tinkering with the zinc coordination site of ReAV.
Front Chem, 12, 2024
|
|
5JZQ
 
 | |
7Z0U
 
 | | Crystal structure of AtWRKY18 DNA-binding domain in complex with W-box DNA | | Descriptor: | DNA (5'-D(*CP*GP*CP*CP*TP*TP*GP*AP*CP*CP*AP*GP*CP*GP*C)-3'), DNA (5'-D(*GP*CP*GP*CP*TP*GP*GP*TP*CP*AP*AP*GP*GP*CP*G)-3'), WRKY transcription factor 18, ... | | Authors: | Grzechowiak, M, Jaskolski, M, Ruszkowski, M. | | Deposit date: | 2022-02-23 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | New aspects of DNA recognition by group II WRKY transcription factor revealed by structural and functional study of AtWRKY18 DNA binding domain.
Int.J.Biol.Macromol., 213, 2022
|
|
7Z0R
 
 | | AtWRKY18 DNA-binding domain | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, NITRATE ION, ... | | Authors: | Grzechowiak, M, Jaskolski, M, Ruszkowski, M. | | Deposit date: | 2022-02-23 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | New aspects of DNA recognition by group II WRKY transcription factor revealed by structural and functional study of AtWRKY18 DNA binding domain.
Int.J.Biol.Macromol., 213, 2022
|
|
1MNO
 
 | | V68N MYOGLOBIN OXY FORM | | Descriptor: | OXYGEN MOLECULE, PROTEIN (MYOGLOBIN), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Murshudov, G.N, Krzywda, S, Brzozowski, A.M, Jaskolski, M, Scott, E.E, Klizas, S.A, Gibson, Q.H, Olson, J.S, Wilkinson, A.J. | | Deposit date: | 1998-08-13 | | Release date: | 1998-08-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Stabilizing bound O2 in myoglobin by valine68 (E11) to asparagine substitution.
Biochemistry, 37, 1998
|
|
6S1U
 
 | | Crystal structure of dimeric M-PMV protease C7A/D26N/C106A mutant in complex with inhibitor | | Descriptor: | Gag-Pro-Pol polyprotein, PRO-0A1-VAL-PSA-ALA-MET-THR | | Authors: | Wosicki, S, Gilski, M, Jaskolski, M, Zabranska, H, Pichova, I. | | Deposit date: | 2019-06-19 | | Release date: | 2019-10-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Comparison of a retroviral protease in monomeric and dimeric states.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6S1W
 
 | | Crystal structure of dimeric M-PMV protease D26N mutant | | Descriptor: | Gag-Pro-Pol polyprotein | | Authors: | Wosicki, S, Gilski, M, Jaskolski, M, Zabranska, H, Pichova, I. | | Deposit date: | 2019-06-19 | | Release date: | 2019-10-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Comparison of a retroviral protease in monomeric and dimeric states.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
1MWC
 
 | | WILD TYPE MYOGLOBIN WITH CO | | Descriptor: | CARBON MONOXIDE, PROTEIN (MYOGLOBIN), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Murshudov, G.N, Krzywda, S, Brzozowski, A.M, Jaskolski, M, Scott, E.E, Klizas, S.A, Gibson, Q.H, Olson, J.S, Wilkinson, A.J. | | Deposit date: | 1998-08-12 | | Release date: | 1998-08-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Stabilizing bound O2 in myoglobin by valine68 (E11) to asparagine substitution.
Biochemistry, 37, 1998
|
|
1MWD
 
 | | WILD TYPE DEOXY MYOGLOBIN | | Descriptor: | PROTEIN (MYOGLOBIN), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Murshudov, G.N, Krzywda, S, Brzozowski, A.M, Jaskolski, M, Scott, E.E, Klizas, A.A, Gibson, Q.H, Olson, J.S, Wilkinson, A.J. | | Deposit date: | 1998-08-11 | | Release date: | 1998-08-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Stabilizing bound O2 in myoglobin by valine68 (E11) to asparagine substitution.
Biochemistry, 37, 1998
|
|
8C23
 
 | | Structure of E. coli Class 2 L-asparaginase EcAIII, mutant M200T (monoclinic form M200T#m) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Sciuk, A, Ruszkowski, M, Jaskolski, M, Loch, J.I. | | Deposit date: | 2022-12-21 | | Release date: | 2023-05-03 | | Last modified: | 2023-06-07 | | Method: | X-RAY DIFFRACTION (1.842 Å) | | Cite: | The effects of nature-inspired amino acid substitutions on structural and biochemical properties of the E. coli L-asparaginase EcAIII.
Protein Sci., 32, 2023
|
|
6S1V
 
 | | Crystal structure of dimeric M-PMV protease D26N mutant in complex with inhibitor | | Descriptor: | Gag-Pro-Pol polyprotein, PRO-0A1-VAL-PSA-ALA-MET-THR | | Authors: | Wosicki, S, Gilski, M, Jaskolski, M, Zabranska, H, Pichova, I. | | Deposit date: | 2019-06-19 | | Release date: | 2019-10-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Comparison of a retroviral protease in monomeric and dimeric states.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
1R4C
 
 | |
2FLH
 
 | | Crystal structure of cytokinin-specific binding protein from mung bean in complex with cytokinin | | Descriptor: | (2E)-2-methyl-4-(9H-purin-6-ylamino)but-2-en-1-ol, SODIUM ION, cytokinin-specific binding protein | | Authors: | Pasternak, O, Bujacz, G.D, Sikorski, M.M, Jaskolski, M. | | Deposit date: | 2006-01-06 | | Release date: | 2006-11-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal Structure of Vigna radiata Cytokinin-Specific Binding Protein in Complex with Zeatin.
Plant Cell, 18, 2006
|
|
5MXB
 
 | | Crystal structure of yellow lupin LLPR-10.2B protein in complex with melatonin | | Descriptor: | Class 10 plant pathogenesis-related protein, N-[2-(5-methoxy-1H-indol-3-yl)ethyl]acetamide, SODIUM ION, ... | | Authors: | Sliwiak, J, Sikorski, M, Jaskolski, M. | | Deposit date: | 2017-01-22 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | PR-10 proteins as potential mediators of melatonin-cytokinin cross-talk in plants: crystallographic studies of LlPR-10.2B isoform from yellow lupine.
FEBS J., 285, 2018
|
|
5MXW
 
 | | Crystal structure of yellow lupin LLPR-10.2B protein in complex with melatonin and trans-zeatin. | | Descriptor: | (2E)-2-methyl-4-(9H-purin-6-ylamino)but-2-en-1-ol, Class 10 plant pathogenesis-related protein, N-[2-(5-methoxy-1H-indol-3-yl)ethyl]acetamide, ... | | Authors: | Sliwiak, J, Sikorski, M, Jaskolski, M. | | Deposit date: | 2017-01-25 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | PR-10 proteins as potential mediators of melatonin-cytokinin cross-talk in plants: crystallographic studies of LlPR-10.2B isoform from yellow lupine.
FEBS J., 285, 2018
|
|
8C0I
 
 | | Structure of E. coli Class 2 L-asparaginase EcAIII, mutant M200L (acyl-enzyme intermediate) | | Descriptor: | CHLORIDE ION, Isoaspartyl peptidase subunit alpha, Isoaspartyl peptidase subunit beta, ... | | Authors: | Sciuk, A, Jaskolski, M, Loch, J.I. | | Deposit date: | 2022-12-16 | | Release date: | 2023-05-03 | | Last modified: | 2023-06-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The effects of nature-inspired amino acid substitutions on structural and biochemical properties of the E. coli L-asparaginase EcAIII.
Protein Sci., 32, 2023
|
|
3E85
 
 | | Crystal Structure of Pathogenesis-related Protein LlPR-10.2B from yellow lupine in complex with Diphenylurea | | Descriptor: | 1,3-DIPHENYLUREA, PR10.2B, SODIUM ION | | Authors: | Fernandes, H.C, Bujacz, G, Bujacz, A, Sikorski, M.M, Jaskolski, M. | | Deposit date: | 2008-08-19 | | Release date: | 2009-03-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Cytokinin-induced structural adaptability of a Lupinus luteus PR-10 protein.
Febs J., 276, 2009
|
|
6QKY
 
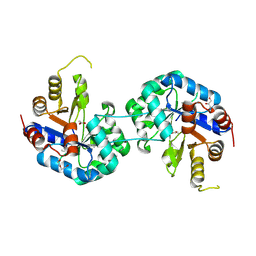 | | Tryptophan synthase subunit alpha from Streptococcus pneumoniae with 3D domain swap in the core of TIM barrel | | Descriptor: | ACETIC ACID, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Michalska, K, Kowiel, M, Bigelow, L, Endres, M, Gilski, M, Jaskolski, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-01-30 | | Release date: | 2019-03-27 | | Last modified: | 2022-03-30 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | 3D domain swapping in the TIM barrel of the alpha subunit of Streptococcus pneumoniae tryptophan synthase.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
4GY9
 
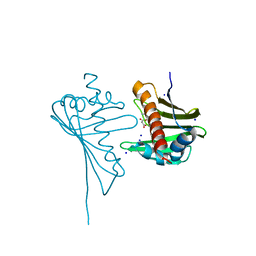 | | Crystal Structure of Medicago truncatula Nodulin 13 (MtN13) in complex with N6-isopentenyladenine (2iP) | | Descriptor: | MALONATE ION, MtN13 protein, N-(3-METHYLBUT-2-EN-1-YL)-9H-PURIN-6-AMINE, ... | | Authors: | Ruszkowski, M, Sikorski, M, Jaskolski, M. | | Deposit date: | 2012-09-05 | | Release date: | 2013-09-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | The landscape of cytokinin binding by a plant nodulin.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
6YEH
 
 | | Arabidopsis thaliana glutamate dehydrogenase isoform 1 in apo form | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Glutamate dehydrogenase 1, POTASSIUM ION | | Authors: | Ruszkowski, M, Grzechowiak, M, Jaskolski, M. | | Deposit date: | 2020-03-24 | | Release date: | 2020-05-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structural Studies of Glutamate Dehydrogenase (Isoform 1) FromArabidopsis thaliana, an Important Enzyme at the Branch-Point Between Carbon and Nitrogen Metabolism.
Front Plant Sci, 11, 2020
|
|
6YEI
 
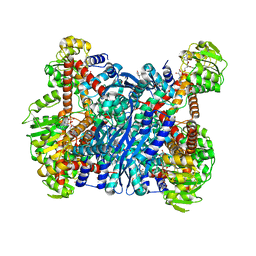 | | Arabidopsis thaliana glutamate dehydrogenase isoform 1 in complex with NAD | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, ... | | Authors: | Ruszkowski, M, Grzechowiak, M, Jaskolski, M. | | Deposit date: | 2020-03-24 | | Release date: | 2020-05-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural Studies of Glutamate Dehydrogenase (Isoform 1) FromArabidopsis thaliana, an Important Enzyme at the Branch-Point Between Carbon and Nitrogen Metabolism.
Front Plant Sci, 11, 2020
|
|
2ZAK
 
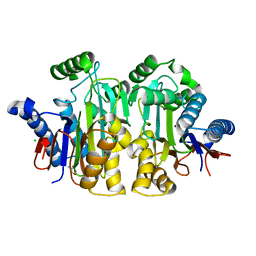 | | Orthorhombic crystal structure of precursor E. coli isoaspartyl peptidase/L-asparaginase (EcAIII) with active-site T179A mutation | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, L-asparaginase precursor, ... | | Authors: | Michalska, K, Hernandez-Santoyo, A, Jaskolski, M. | | Deposit date: | 2007-10-07 | | Release date: | 2008-03-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal packing of plant-type L-asparaginase from Escherichia coli
Acta Crystallogr.,Sect.D, 64, 2008
|
|
4PV3
 
 | | Crystal structure of potassium-dependent plant-type L-asparaginase from Phaseolus vulgaris in complex with Na+ cations | | Descriptor: | L-ASPARAGINASE ALPHA SUBUNIT, L-ASPARAGINASE BETA SUBUNIT, SODIUM ION | | Authors: | Bejger, M, Gilski, M, Imiolczyk, B, Jaskolski, M. | | Deposit date: | 2014-03-14 | | Release date: | 2014-09-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Na+/K+ exchange switches the catalytic apparatus of potassium-dependent plant L-asparaginase
Acta Crystallogr.,Sect.D, 70, 2014
|
|
