6MFJ
 
 | |
4L4W
 
 | |
4KW5
 
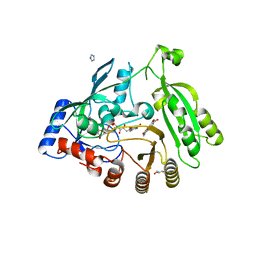 | | M. tuberculosis DprE1 in complex with inhibitor TCA1 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, IMIDAZOLE, ... | | Authors: | Batt, S.M, Besra, G.S, Futterer, K. | | Deposit date: | 2013-05-23 | | Release date: | 2013-07-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.612 Å) | | Cite: | Identification of a small molecule with activity against drug-resistant and persistent tuberculosis.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4KVN
 
 | | Crystal structure of Fab 39.29 in complex with Influenza Hemagglutinin A/Perth/16/2009 (H3N2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | Authors: | Fong, R, Swem, L.R, Lupardus, P.J. | | Deposit date: | 2013-05-22 | | Release date: | 2013-08-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A Novel In vivo Human Plasmablast Enrichment Technique Allows Rapid Identification of Therapeutic Anti-Influenza A Antibodies
Cell Host Microbe, 14, 2013
|
|
5E8B
 
 | |
5EMX
 
 | |
4O9C
 
 | | Crystal structure of Beta-ketothiolase (PhaA) from Ralstonia eutropha H16 | | Descriptor: | Acetyl-CoA acetyltransferase, COENZYME A | | Authors: | Kim, E.J, Kim, J, Kim, S, Kim, K.J. | | Deposit date: | 2014-01-02 | | Release date: | 2014-12-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure and biochemical characterization of PhaA from Ralstonia eutropha, a polyhydroxyalkanoate-producing bacterium.
Biochem.Biophys.Res.Commun., 452, 2014
|
|
4O9A
 
 | | Crystal structure of Beta-ketothiolase (PhaA) from Ralstonia eutropha H16 | | Descriptor: | Acetyl-CoA acetyltransferase | | Authors: | Kim, E.J, Kim, J, Kim, S, Kim, K.J. | | Deposit date: | 2014-01-02 | | Release date: | 2015-02-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Crystal structure and biochemical characterization of PhaA from Ralstonia eutropha, a polyhydroxyalkanoate-producing bacterium.
Biochem.Biophys.Res.Commun., 452, 2014
|
|
4O99
 
 | | Crystal structure of Beta-ketothiolase (PhaA) from Ralstonia eutropha H16 | | Descriptor: | Acetyl-CoA acetyltransferase, GLYCEROL | | Authors: | Kim, E.J, Kim, J, Kim, S, Kim, K.J. | | Deposit date: | 2014-01-02 | | Release date: | 2014-12-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal structure and biochemical characterization of PhaA from Ralstonia eutropha, a polyhydroxyalkanoate-producing bacterium.
Biochem.Biophys.Res.Commun., 452, 2014
|
|
6JN8
 
 | | Structure of H216A mutant open form peptidoglycan peptidase | | Descriptor: | Peptidase M23, SULFATE ION, ZINC ION | | Authors: | Min, K.J, An, D.R, Yoon, H.J, Suh, S.W, Lee, H.H. | | Deposit date: | 2019-03-13 | | Release date: | 2020-01-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.106 Å) | | Cite: | Peptidoglycan reshaping by a noncanonical peptidase for helical cell shape in Campylobacter jejuni.
Nat Commun, 11, 2020
|
|
6JMZ
 
 | | Structure of H247A mutant open form peptidoglycan peptidase | | Descriptor: | Peptidase M23, ZINC ION | | Authors: | Min, K.J, An, D.R, Yoon, H.J, Suh, S.W, Lee, H.H. | | Deposit date: | 2019-03-13 | | Release date: | 2020-01-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Peptidoglycan reshaping by a noncanonical peptidase for helical cell shape in Campylobacter jejuni.
Nat Commun, 11, 2020
|
|
6JMX
 
 | | Structure of open form of peptidoglycan peptidase | | Descriptor: | D(-)-TARTARIC ACID, GLYCEROL, Peptidase M23, ... | | Authors: | Min, K.J, An, D.R, Yoon, H.J, Suh, S.W, Lee, H.H. | | Deposit date: | 2019-03-13 | | Release date: | 2020-01-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.859 Å) | | Cite: | Peptidoglycan reshaping by a noncanonical peptidase for helical cell shape in Campylobacter jejuni.
Nat Commun, 11, 2020
|
|
6JN1
 
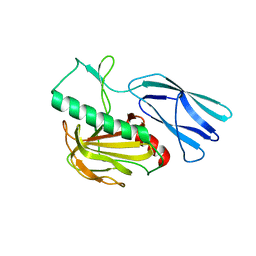 | | Structure of H247A mutant peptidoglycan peptidase complex with penta peptide | | Descriptor: | C0O-DAL-DAL, Peptidase M23, ZINC ION | | Authors: | Min, K.J, An, D.R, Yoon, H.J, Suh, S.W, Lee, H.H. | | Deposit date: | 2019-03-13 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.382 Å) | | Cite: | Peptidoglycan reshaping by a noncanonical peptidase for helical cell shape in Campylobacter jejuni.
Nat Commun, 11, 2020
|
|
6JN0
 
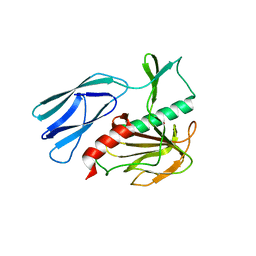 | | Structure of H247A mutant peptidoglycan peptidase complex with tetra-tri peptide | | Descriptor: | C0O-DAL-API, Peptidase M23, ZINC ION | | Authors: | Min, K.J, An, D.R, Yoon, H.J, Suh, S.W, Lee, H.H. | | Deposit date: | 2019-03-13 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.164 Å) | | Cite: | Peptidoglycan reshaping by a noncanonical peptidase for helical cell shape in Campylobacter jejuni.
Nat Commun, 11, 2020
|
|
6JN7
 
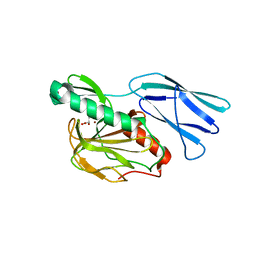 | | Structure of H216A mutant closed form peptidoglycan peptidase | | Descriptor: | D(-)-TARTARIC ACID, Peptidase M23, ZINC ION | | Authors: | Min, K.J, An, D.R, Yoon, H.J, Suh, S.W, Lee, H.H. | | Deposit date: | 2019-03-13 | | Release date: | 2020-01-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Peptidoglycan reshaping by a noncanonical peptidase for helical cell shape in Campylobacter jejuni.
Nat Commun, 11, 2020
|
|
6JMY
 
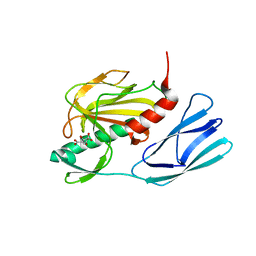 | | Structure of wild type closed form of peptidoglycan peptidase | | Descriptor: | CITRIC ACID, Peptidase M23, ZINC ION | | Authors: | Min, K.J, An, D.R, Yoon, H.J, Suh, S.W, Lee, H.H. | | Deposit date: | 2019-03-13 | | Release date: | 2020-01-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.661 Å) | | Cite: | Peptidoglycan reshaping by a noncanonical peptidase for helical cell shape in Campylobacter jejuni.
Nat Commun, 11, 2020
|
|
6KV1
 
 | | Structure of wild type closed form of peptidoglycan peptidase ZN SAD | | Descriptor: | CITRIC ACID, Peptidase M23, ZINC ION | | Authors: | Min, K.J, An, D.R, Yoon, H.J, Suh, S.W, Lee, H.H. | | Deposit date: | 2019-09-03 | | Release date: | 2020-01-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.722 Å) | | Cite: | Peptidoglycan reshaping by a noncanonical peptidase for helical cell shape in Campylobacter jejuni.
Nat Commun, 11, 2020
|
|
4DS2
 
 | |
7WZE
 
 | |
4G68
 
 | |
7WKR
 
 | |
7WUC
 
 | |
7XUG
 
 | |
7XUE
 
 | |
7XUI
 
 | |
