2PDH
 
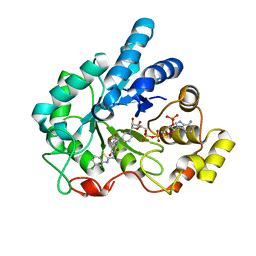 | | Human aldose reductase mutant L300P complexed with uracil-type inhibitor at 1.45 A. | | Descriptor: | Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, {3-[(5-CHLORO-1,3-BENZOTHIAZOL-2-YL)METHYL]-2,4-DIOXO-3,4-DIHYDROPYRIMIDIN-1(2H)-YL}ACETIC ACID | | Authors: | Steuber, H, Heine, A, Klebe, G. | | Deposit date: | 2007-03-31 | | Release date: | 2008-04-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Merging the binding sites of aldose and aldehyde reductase for detection of inhibitor selectivity-determining features.
J.Mol.Biol., 379, 2008
|
|
2D23
 
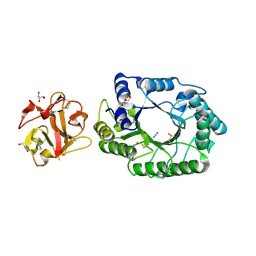 | | Crystal structure of EP complex of catalytic-site mutant xylanase from Streptomyces olivaceoviridis E-86 | | Descriptor: | AZIDE ION, ENDO-1,4-BETA-D-XYLANASE, GLYCEROL, ... | | Authors: | Suzuki, R, Kuno, A, Fujimoto, Z, Ito, S, Kawahara, S.I, Kaneko, S, Hasegawa, T, Taira, K. | | Deposit date: | 2005-09-02 | | Release date: | 2006-10-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystallographic snapshots of an entire reaction cycle for a retaining xylanase from Streptomyces olivaceoviridis E-86
J.Biochem., 146, 2009
|
|
1K7C
 
 | |
4CKY
 
 | | Structure of the Mycobacterium tuberculosis Type II Dehydroquinase inhibited by a 3-dehydroquinic acid derivative | | Descriptor: | 2,2-dimethyl-3-dehydroquinic acid, 3-DEHYDROQUINATE DEHYDRATASE, SODIUM ION, ... | | Authors: | Otero, J.M, Llamas-Saiz, A.L, Maneiro, M, Peon, A, Sedes, A, Lamb, H, Hawkins, A.R, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2014-01-10 | | Release date: | 2015-03-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Investigation of the Dehydratation Mechanism Catalyzed by the Type II Dehydroquinase
To be Published
|
|
1K9F
 
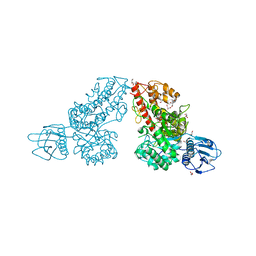 | | Crystal structure of a mutated family-67 alpha-D-glucuronidase (E285N) from Bacillus stearothermophilus T-6, complexed with aldotetraouronic acid | | Descriptor: | 4-O-methyl-alpha-D-glucopyranuronic acid-(1-2)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, GLYCEROL, alpha-D-glucuronidase | | Authors: | Golan, G, Shallom, D, Teplitsky, A, Zaide, G, Shulami, S, Baasov, T, Stojanoff, V, Thompson, A, Shoham, Y, Shoham, G. | | Deposit date: | 2001-10-29 | | Release date: | 2002-10-29 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structures of Geobacillus stearothermophilus {alpha}-Glucuronidase Complexed with Its Substrate and Products: MECHANISTIC IMPLICATIONS.
J.Biol.Chem., 279, 2004
|
|
1K5N
 
 | | HLA-B*2709 BOUND TO NONA-PEPTIDE M9 | | Descriptor: | GLYCEROL, beta-2-microglobulin, light chain, ... | | Authors: | Hulsmeyer, M, Hillig, R.C, Volz, A, Ruhl, M, Schroder, W, Saenger, W, Ziegler, A, Uchanska-Ziegler, B. | | Deposit date: | 2001-10-11 | | Release date: | 2002-10-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | HLA-B27 Subtypes Differentially Associated with Disease Exhibit Subtle Structural Alterations
J.Biol.Chem., 277, 2002
|
|
2D1Z
 
 | | Crystal structure of catalytic-site mutant xylanase from Streptomyces olivaceoviridis E-86 | | Descriptor: | ENDO-1,4-BETA-D-XYLANASE, GLYCEROL, SULFATE ION | | Authors: | Suzuki, R, Kuno, A, Fujimoto, Z, Ito, S, Kawahara, S.I, Kaneko, S, Hasegawa, T, Taira, K. | | Deposit date: | 2005-09-02 | | Release date: | 2006-10-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallographic snapshots of an entire reaction cycle for a retaining xylanase from Streptomyces olivaceoviridis E-86
J.Biochem., 146, 2009
|
|
1K77
 
 | | Crystal Structure of EC1530, a Putative Oxygenase from Escherichia coli | | Descriptor: | FORMIC ACID, GLYCEROL, Hypothetical protein ygbM, ... | | Authors: | Kim, Y, Skarina, T, Beasley, S, Laskowski, R, Arrowsmith, C.H, Joachimiak, A, Edwards, A.M, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2001-10-18 | | Release date: | 2002-03-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystal structure of Escherichia coli EC1530, a glyoxylate induced protein YgbM.
Proteins, 48, 2002
|
|
6ZAB
 
 | | Structure of the transcriptional repressor Atu1419 (VanR) from agrobacterium fabrum in complex a palindromic DNA (P6422 space group) | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Morera, S, Naretto, A, Vigouroux, A, Legrand, P. | | Deposit date: | 2020-06-05 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Characterization of the first tetrameric transcription factor of the GntR superfamily with allosteric regulation from the bacterial pathogen Agrobacterium fabrum.
Nucleic Acids Res., 49, 2021
|
|
8B30
 
 | |
2D20
 
 | | Crystal structure of michaelis complex of catalytic-site mutant xylanase from Streptomyces olivaceoviridis E-86 | | Descriptor: | ENDO-1,4-BETA-D-XYLANASE, GLYCEROL, P-NITROPHENOL, ... | | Authors: | Suzuki, R, Kuno, A, Fujimoto, Z, Ito, S, Kawahara, S.I, Kaneko, S, Hasegawa, T, Taira, K. | | Deposit date: | 2005-09-02 | | Release date: | 2006-10-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystallographic snapshots of an entire reaction cycle for a retaining xylanase from Streptomyces olivaceoviridis E-86
J.Biochem., 146, 2009
|
|
4CKZ
 
 | | Structure of the Mycobacterium tuberculosis Type II Dehydroquinase D88N mutant | | Descriptor: | 3-DEHYDROQUINATE DEHYDRATASE | | Authors: | Otero, J.M, Llamas-Saiz, A.L, Maneiro, M, Peon, A, Sedes, A, Lamb, H, Hawkins, A.R, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2014-01-10 | | Release date: | 2015-03-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Investigation of the Dehydratation Mechanism Catalyzed by the Type II Dehydroquinase
To be Published
|
|
2OXB
 
 | | Crystal structure of a cell-wall invertase (E203Q) from Arabidopsis thaliana in complex with sucrose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-fructofuranosidase, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Lammens, W, Le Roy, K, Van Laere, A, Van den Ende, W, Rabijns, A. | | Deposit date: | 2007-02-20 | | Release date: | 2008-01-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | An alternate sucrose binding mode in the E203Q Arabidopsis invertase mutant: An X-ray crystallography and docking study.
Proteins, 71, 2007
|
|
2DD6
 
 | | Solution structure of Dermaseptin antimicrobial peptide truncated, mutated analog, K4-S4(1-13)a | | Descriptor: | Dermaseptin-4 | | Authors: | Shalev, D.E, Rotem, S, Fish, A, Mor, A. | | Deposit date: | 2006-01-19 | | Release date: | 2006-02-28 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Consequences of N-acylation on structure and membrane binding properties of dermaseptin derivative k4-s4-(1-13)
J.Biol.Chem., 281, 2006
|
|
1K9E
 
 | | Crystal structure of a mutated family-67 alpha-D-glucuronidase (E285N) from Bacillus stearothermophilus T-6, complexed with 4-O-methyl-glucuronic acid | | Descriptor: | 4-O-methyl-alpha-D-glucopyranuronic acid, GLYCEROL, alpha-D-glucuronidase | | Authors: | Golan, G, Shallom, D, Teplitsky, A, Zaide, G, Shulami, S, Baasov, T, Stojanoff, V, Thompson, A, Shoham, Y, Shoham, G. | | Deposit date: | 2001-10-29 | | Release date: | 2002-10-29 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structures of Geobacillus stearothermophilus {alpha}-Glucuronidase Complexed with Its Substrate and Products: MECHANISTIC IMPLICATIONS.
J.Biol.Chem., 279, 2004
|
|
6EON
 
 | | Galactanase BT0290 | | Descriptor: | Beta-galactosidase, CALCIUM ION, alpha-D-galactopyranose | | Authors: | Basle, A, Munoz, J, Gilbert, H. | | Deposit date: | 2017-10-10 | | Release date: | 2017-11-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A surface endogalactanase in Bacteroides thetaiotaomicron confers keystone status for arabinogalactan degradation.
Nat Microbiol, 3, 2018
|
|
4CZU
 
 | |
4D28
 
 | |
1K9D
 
 | | The 1.7 A crystal structure of alpha-D-glucuronidase, a family-67 glycoside hydrolase from Bacillus stearothermophilus T-1 | | Descriptor: | GLYCEROL, alpha-D-glucuronidase | | Authors: | Golan, G, Shallom, D, Teplitsky, A, Zaide, G, Shulami, S, Baasov, T, Stojanoff, V, Thompson, A, Shoham, Y, Shoham, G. | | Deposit date: | 2001-10-29 | | Release date: | 2002-10-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structures of Geobacillus stearothermophilus {alpha}-Glucuronidase Complexed with Its Substrate and Products: MECHANISTIC IMPLICATIONS.
J.Biol.Chem., 279, 2004
|
|
1K3R
 
 | | Crystal Structure of the Methyltransferase with a Knot from Methanobacterium thermoautotrophicum | | Descriptor: | conserved protein MT0001 | | Authors: | Zarembinski, T.I, Kim, Y, Peterson, K, Christendat, D, Dharamsi, A, Arrowsmith, C.H, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2001-10-03 | | Release date: | 2002-05-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Deep trefoil knot implicated in RNA binding found in an archaebacterial protein.
Proteins, 50, 2003
|
|
2DH1
 
 | | Crystal structure of peanut lectin lactose-azobenzene-4,4'-dicarboxylic acid-lactose complex | | Descriptor: | Galactose-binding lectin | | Authors: | Natchiar, S.K, Srinivas, O, Nivedita, M, Sagarika, D, Jayaraman, N, Surolia, A, Vijayan, M. | | Deposit date: | 2006-03-17 | | Release date: | 2006-08-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (7.65 Å) | | Cite: | Multivalency in lectins - A crystallographic, modelling and light-scattering study involving peanut lectin and a bivalent ligand
Curr.Sci., 90, 2006
|
|
5B1L
 
 | | The mouse nucleosome structure containing H3t | | Descriptor: | CHLORIDE ION, DNA (146-MER), Histone H2A type 1, ... | | Authors: | Urahama, T, Machida, S, Horikoshi, N, Osakabe, A, Tachiwana, H, Taguchi, H, Kurumizaka, H. | | Deposit date: | 2015-12-08 | | Release date: | 2017-02-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Testis-Specific Histone Variant H3t Gene Is Essential for Entry into Spermatogenesis
Cell Rep, 18, 2017
|
|
6QMM
 
 | | Crystal structure of Synecochoccus Spermidine Synthase in complex with putrescine, spermidine and MTA | | Descriptor: | (2S)-1-[3-{[(2R)-2-hydroxypropyl]oxy}-2,2-bis({[(2R)-2-hydroxypropyl]oxy}methyl)propoxy]propan-2-ol, 1,4-DIAMINOBUTANE, 5'-DEOXY-5'-METHYLTHIOADENOSINE, ... | | Authors: | Guedez, G, Pothipongsa, A, Incharoensakdi, A, Salminen, T.A. | | Deposit date: | 2019-02-07 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Crystal structure of dimericSynechococcusspermidine synthase with bound polyamine substrate and product.
Biochem.J., 476, 2019
|
|
6QPA
 
 | | Halothiobacillus neapolitanus sulfur oxygenase reductase | | Descriptor: | FE (III) ION, SULFATE ION, Sulfur oxygenase/reductase | | Authors: | Frazao, C, Klezin, A, Poell, U, Veith, A, Ruehl, P, Coelho, R. | | Deposit date: | 2019-02-13 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Multiple sulfane modifications in active-site cysteine thiols of two sulfur oxygenase reductases and analysis of substrate/product channels
To Be Published
|
|
4COS
 
 | | Crystal structure of the PHD-Bromo-PWWP cassette of human PRKCBP1 | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, PROTEIN KINASE C-BINDING PROTEIN 1, ZINC ION | | Authors: | Krojer, T, Savitsky, P, Newman, J.A, Cooper, C.D.O, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A, Filippakopoulos, P. | | Deposit date: | 2014-01-30 | | Release date: | 2014-03-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Multivalent Histone and DNA Engagement by a PHD/BRD/PWWP Triple Reader Cassette Recruits ZMYND8 to K14ac-Rich Chromatin.
Cell Rep, 17, 2016
|
|
