7F0I
 
 | | phosphodiesterase-9A in complex with inhibitor 4b | | Descriptor: | 1-cyclopentyl-6-[[(2R)-1-(6-fluoranyl-2-azaspiro[3.3]heptan-2-yl)-1-oxidanylidene-propan-2-yl]amino]-5H-pyrazolo[3,4-d]pyrimidin-4-one, Isoform PDE9A2 of High affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A, MAGNESIUM ION, ... | | Authors: | Wu, Y, Huang, Y.Y, Luo, H.B. | | Deposit date: | 2021-06-04 | | Release date: | 2022-04-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.70000887 Å) | | Cite: | Discovery of Potent Phosphodiesterase-9 Inhibitors for the Treatment of Hepatic Fibrosis
J.Med.Chem., 64, 2021
|
|
7F1E
 
 | | Structure of METTL6 bound with SAM | | Descriptor: | S-ADENOSYLMETHIONINE, tRNA N(3)-methylcytidine methyltransferase METTL6 | | Authors: | Li, S, Liao, S, Xu, C. | | Deposit date: | 2021-06-09 | | Release date: | 2022-01-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.589 Å) | | Cite: | Structural basis for METTL6-mediated m3C RNA methylation.
Biochem.Biophys.Res.Commun., 589, 2021
|
|
7D2Y
 
 | | complex of two RRM domains | | Descriptor: | Embryonic developmental protein tofu-6, RRM2, SULFATE ION | | Authors: | Wang, X, Liao, S, Xu, C. | | Deposit date: | 2020-09-17 | | Release date: | 2021-08-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Molecular basis for PICS-mediated piRNA biogenesis and cell division.
Nat Commun, 12, 2021
|
|
7CNG
 
 | | Structure of CDK5R1 bound FEM1B | | Descriptor: | Protein fem-1 homolog B,Peptide from Cyclin-dependent kinase 5 activator 1, SULFATE ION | | Authors: | Chen, X, Liao, S, Xu, C. | | Deposit date: | 2020-07-31 | | Release date: | 2020-10-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.49 Å) | | Cite: | Molecular basis for arginine C-terminal degron recognition by Cul2 FEM1 E3 ligase.
Nat.Chem.Biol., 17, 2021
|
|
7EJS
 
 | | Structure of ERH-2 bound to PICS-1 | | Descriptor: | Enhancer of rudimentary homolog 2,Protein pid-3 | | Authors: | Wang, X, Xu, C. | | Deposit date: | 2021-04-02 | | Release date: | 2021-08-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.387 Å) | | Cite: | Molecular basis for PICS-mediated piRNA biogenesis and cell division.
Nat Commun, 12, 2021
|
|
7EJO
 
 | | Structure of ERH-2 bound to TOST-1 | | Descriptor: | Enhancer of rudimentary homolog 2, Enhancer of rudimentary homolog 2,Protein tost-1 | | Authors: | Wang, X, Xu, C. | | Deposit date: | 2021-04-02 | | Release date: | 2021-08-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.191 Å) | | Cite: | Molecular basis for PICS-mediated piRNA biogenesis and cell division.
Nat Commun, 12, 2021
|
|
7FAW
 
 | | Structure of LW domain from Yeast | | Descriptor: | Transcription elongation factor S-II | | Authors: | Liao, S, Gao, J, Tu, X. | | Deposit date: | 2021-07-07 | | Release date: | 2022-07-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.438 Å) | | Cite: | Structural basis for evolutionarily conserved interactions between TFIIS and Paf1C.
Int.J.Biol.Macromol., 253, 2023
|
|
7VP9
 
 | | Crystal structure of human ClpP in complex with ZG111 | | Descriptor: | (6S,9aS)-N-[(4-bromophenyl)methyl]-6-[(2S)-butan-2-yl]-8-(naphthalen-1-ylmethyl)-4,7-bis(oxidanylidene)-3,6,9,9a-tetrahydro-2H-pyrazino[1,2-a]pyrimidine-1-carboxamide, ATP-dependent Clp protease proteolytic subunit, mitochondrial, ... | | Authors: | Wang, P.Y, Gan, J.H, Yang, C.-G. | | Deposit date: | 2021-10-15 | | Release date: | 2022-06-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.552 Å) | | Cite: | Aberrant human ClpP activation disturbs mitochondrial proteome homeostasis to suppress pancreatic ductal adenocarcinoma.
Cell Chem Biol, 29, 2022
|
|
7XQ5
 
 | | Crystal structure of ScIno2p-ScIno4p bound promoter DNA | | Descriptor: | DNA (5'-D(*GP*AP*TP*TP*TP*TP*CP*AP*CP*AP*TP*GP*CP*AP*G)-3'), DNA (5'-D(P*CP*CP*TP*GP*CP*AP*TP*GP*TP*GP*AP*AP*AP*AP*T)-3'), HEXANE-1,6-DIOL, ... | | Authors: | Khan, M.H, Lu, X. | | Deposit date: | 2022-05-06 | | Release date: | 2022-07-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Analysis of Ino2p/Ino4p Mutual Interactions and Their Binding Interface with Promoter DNA.
Int J Mol Sci, 23, 2022
|
|
7X39
 
 | | Structure of CIZ1 bound ERH | | Descriptor: | Enhancer of rudimentary homolog,Cip1-interacting zinc finger protein | | Authors: | Wang, X, Xu, C. | | Deposit date: | 2022-02-28 | | Release date: | 2022-08-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Molecular basis for the recognition of CIZ1 by ERH.
Febs J., 290, 2023
|
|
7V5Y
 
 | |
7V5Z
 
 | |
7V6W
 
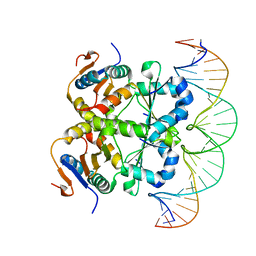 | | Crystal structure of heterohexameric Sa2YoeB-Sa2YefM complex bound to 26bp-DNA | | Descriptor: | Antitoxin, DNA (25-MER), DNA (26-MER), ... | | Authors: | Xue, L, Khan, M.H, Yue, J. | | Deposit date: | 2021-08-20 | | Release date: | 2021-12-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The two paralogous copies of the YoeB-YefM toxin-antitoxin module in Staphylococcus aureus differ in DNA binding and recognition patterns.
J.Biol.Chem., 298, 2022
|
|
7XGW
 
 | |
7YCV
 
 | |
7YCU
 
 | |
7YCW
 
 | |
7YCS
 
 | |
7YF2
 
 | |
7Y9C
 
 | |
7YF4
 
 | |
7YF3
 
 | |
2GVF
 
 | | HCV NS3-4A protease domain complexed with a macrocyclic ketoamide inhibitor, SCH419021 | | Descriptor: | (6R,8S,11S)-11-CYCLOHEXYL-N-(1-{[(2-{[(1S)-2-(DIMETHYLAMINO)-2-OXO-1-PHENYLETHYL]AMINO}-2-OXOETHYL)AMINO](OXO)ACETYL}BUTYL)-10,13-DIOXO-2,5-DIOXA-9,12-DIAZATRICYCLO[14.3.1.1~6,9~]HENICOSA-1(20),16,18-TRIENE-8-CARBOXAMIDE, Polyprotein, ZINC ION, ... | | Authors: | Arasappan, A, Njoroge, F.G, Chen, K.X, Venkatraman, S, Parekh, T.N, Gu, H, Pichardo, J, Butkiewicz, N, Prongay, A, Madison, V, Girijavallabhan, V. | | Deposit date: | 2006-05-02 | | Release date: | 2007-01-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | P2-P4 macrocyclic inhibitors of hepatitis C virus NS3-4A serine protease.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
