4C9Z
 
 | | Crystal structure of Siah1 at 1.95 A resolution | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, E3 UBIQUITIN-PROTEIN LIGASE SIAH1, ... | | Authors: | Rimsa, V, Eadsforth, T.C, Hunter, W.N. | | Deposit date: | 2013-10-04 | | Release date: | 2013-10-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Two High-Resolution Structures of the Human E3 Ubiquitin Ligase Siah1.
Acta Crystallogr.,Sect.F, 96, 2013
|
|
4C81
 
 | | IspF (Plasmodium falciparum) CDP complex | | Descriptor: | 2-C-METHYL-D-ERYTHRITOL 2,4-CYCLODIPHOSPHATE SYNTHASE, CHLORIDE ION, CYTIDINE-5'-DIPHOSPHATE, ... | | Authors: | O'Rourke, P.E.F, Kalinowska-Tluscik, J, Fyfe, P.K, Dawson, A, Hunter, W.N. | | Deposit date: | 2013-09-27 | | Release date: | 2014-01-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Crystal Structures of Ispf from Plasmodium Falciparum and Burkholderia Cenocepacia: Comparisons Inform Antimicrobial Drug Target Assessment.
Bmc Struct.Biol., 14, 2014
|
|
4C82
 
 | | IspF (Plasmodium falciparum) unliganded structure | | Descriptor: | 2-C-METHYL-D-ERYTHRITOL 2,4-CYCLODIPHOSPHATE SYNTHASE, SULFATE ION, ZINC ION | | Authors: | O'Rourke, P.E.F, Kalinowska-Tluscik, J, Fyfe, P.K, Dawson, A, Hunter, W.N. | | Deposit date: | 2013-09-27 | | Release date: | 2014-01-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of Ispf from Plasmodium Falciparum and Burkholderia Cenocepacia: Comparisons Inform Antimicrobial Drug Target Assessment.
Bmc Struct.Biol., 14, 2014
|
|
4C8E
 
 | | IspF (Burkholderia cenocepacia) 2CMP complex | | Descriptor: | 2-C-METHYL-D-ERYTHRITOL 2,4-CYCLODIPHOSPHATE SYNTHASE, CYTIDINE-5'-MONOPHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | O'Rourke, P.E.F, Kalinowska-Tluscik, J, Fyfe, P.K, Dawson, A, Hunter, W.N. | | Deposit date: | 2013-09-30 | | Release date: | 2014-01-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structures of Ispf from Plasmodium Falciparum and Burkholderia Cenocepacia: Comparisons Inform Antimicrobial Drug Target Assessment.
Bmc Struct.Biol., 14, 2014
|
|
4B4W
 
 | | Crystal structure of Acinetobacter baumannii N5, N10- methylenetetrahydrofolate dehydrogenase-cyclohydrolase (FolD) complexed with NADP cofactor and an inhibitor | | Descriptor: | (2S)-2-[[4-[[2,4-bis(azanyl)-6-oxidanylidene-1H-pyrimidin-5-yl]carbamoylamino]phenyl]carbonylamino]pentanedioic acid, BIFUNCTIONAL PROTEIN FOLD, CHLORIDE ION, ... | | Authors: | Eadsforth, T.C, Maluf, F.V, Hunter, W.N. | | Deposit date: | 2012-08-01 | | Release date: | 2012-08-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Acinetobacter Baumannii Fold Ligand Complexes; Potent Inhibitors of Folate Metabolism and a Re-Evaluation of the Ly374571 Structure.
FEBS J., 279, 2012
|
|
4B4U
 
 | | Crystal structure of Acinetobacter baumannii N5, N10- methylenetetrahydrofolate dehydrogenase-cyclohydrolase (FolD) complexed with NADP cofactor | | Descriptor: | 1,2-ETHANEDIOL, BIFUNCTIONAL PROTEIN FOLD, CHLORIDE ION, ... | | Authors: | Eadsforth, T.C, Maluf, F.V, Hunter, W.N. | | Deposit date: | 2012-08-01 | | Release date: | 2012-08-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Acinetobacter Baumannii Fold Ligand Complexes; Potent Inhibitors of Folate Metabolism and a Re-Evaluation of the Ly374571 Structure.
FEBS J., 279, 2012
|
|
4CLX
 
 | |
4CM1
 
 | | Crystal structure of pteridine reductase 1 (PTR1) from Trypanosoma brucei in ternary complex with cofactor and inhibitor | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 5-(p-tolyl)-7H-pyrrolo[2,3-d]pyrimidine-2,4-diamine, ACETATE ION, ... | | Authors: | Barrack, K.L, Hunter, W.N. | | Deposit date: | 2014-01-15 | | Release date: | 2015-01-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Based Design and Synthesis of Antiparasitic Pyrrolopyrimidines Targeting Pteridine Reductase 1.
J.Med.Chem., 57, 2014
|
|
4WCD
 
 | | Trypanosoma brucei PTR1 in complex with inhibitor 10 | | Descriptor: | 5-(1H-benzotriazol-6-yl)-1,3,4-thiadiazol-2-amine, ACETATE ION, GLYCEROL, ... | | Authors: | Mangani, S, Di Pisa, F, Pozzi, C. | | Deposit date: | 2014-09-04 | | Release date: | 2015-09-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Exploiting the 2-Amino-1,3,4-thiadiazole Scaffold To Inhibit Trypanosoma brucei Pteridine Reductase in Support of Early-Stage Drug Discovery.
ACS Omega, 2, 2017
|
|
4WCF
 
 | | Trypanosoma brucei PTR1 in complex with inhibitor 9 | | Descriptor: | 3-(5-amino-1,3,4-thiadiazol-2-yl)pyridin-4-amine, ACETATE ION, GLYCEROL, ... | | Authors: | Mangani, S, Di Pisa, F, Pozzi, C. | | Deposit date: | 2014-09-04 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Exploiting the 2-Amino-1,3,4-thiadiazole Scaffold To Inhibit Trypanosoma brucei Pteridine Reductase in Support of Early-Stage Drug Discovery.
ACS Omega, 2, 2017
|
|
2WD8
 
 | | PTERIDINE REDUCTASE 1 (PTR1) FROM TRYPANOSOMA BRUCEI IN COMPLEX WITH NADP AND DDD00071204 | | Descriptor: | 1-(3,4-DICHLOROBENZYL)-7-PHENYL-1H-BENZIMIDAZOL-2-AMINE, CHLORIDE ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Robinson, D.A, Wyatt, P.G, Spinks, D, Brenk, R. | | Deposit date: | 2009-03-20 | | Release date: | 2009-06-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | One Scaffold, Three Binding Modes: Novel and Selective Pteridine Reductase 1 Inhibitors Derived from Fragment Hits Discovered by Virtual Screening.
J.Med.Chem., 52, 2009
|
|
3OC2
 
 | | Crystal structure of penicillin-binding protein 3 from Pseudomonas aeruginosa | | Descriptor: | CHLORIDE ION, Penicillin-binding protein 3 | | Authors: | Sainsbury, S, Bird, L, Stuart, D.I, Owens, R.J, Ren, J, Oxford Protein Production Facility (OPPF) | | Deposit date: | 2010-08-09 | | Release date: | 2010-11-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.968 Å) | | Cite: | Crystal structures of penicillin-binding protein 3 from Pseudomonas aeruginosa: comparison of native and antibiotic-bound forms
J.Mol.Biol., 405, 2011
|
|
3OCN
 
 | | Crystal structure of penicillin-binding protein 3 from Pseudomonas aeruginosa in complex with ceftazidime | | Descriptor: | 1-({(2R)-2-[(1R)-1-{[(2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-{[(2-carboxypropan-2-yl)oxy]imino}acetyl]amino}-2-oxoethyl]-4-carboxy-3,6-dihydro-2H-1,3-thiazin-5-yl}methyl)pyridinium, penicillin-binding protein 3 | | Authors: | Sainsbury, S, Bird, L, Stuart, D.I, Owens, R.J, Ren, J, Oxford Protein Production Facility (OPPF) | | Deposit date: | 2010-08-10 | | Release date: | 2010-11-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Crystal structures of penicillin-binding protein 3 from Pseudomonas aeruginosa: comparison of native and antibiotic-bound forms
J.Mol.Biol., 405, 2011
|
|
3OCL
 
 | | Crystal structure of penicillin-binding protein 3 from Pseudomonas aeruginosa in complex with carbenicillin | | Descriptor: | (2R,4S)-2-[(1R)-1-{[(2S)-2-carboxy-2-phenylacetyl]amino}-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Sainsbury, S, Bird, L, Stuart, D.I, Owens, R.J, Ren, J, Oxford Protein Production Facility (OPPF) | | Deposit date: | 2010-08-10 | | Release date: | 2010-11-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of penicillin-binding protein 3 from Pseudomonas aeruginosa: comparison of native and antibiotic-bound forms
J.Mol.Biol., 405, 2011
|
|
1AH5
 
 | | REDUCED FORM SELENOMETHIONINE-LABELLED HYDROXYMETHYLBILANE SYNTHASE DETERMINED BY MAD | | Descriptor: | 3-[5-{[3-(2-carboxyethyl)-4-(carboxymethyl)-5-methyl-1H-pyrrol-2-yl]methyl}-4-(carboxymethyl)-1H-pyrrol-3-yl]propanoic acid, HYDROXYMETHYLBILANE SYNTHASE | | Authors: | Helliwell, J.R, Nieh, Y.P, Harrop, S.J, Cassetta, A. | | Deposit date: | 1997-04-13 | | Release date: | 1997-10-15 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Determination of the structure of seleno-methionine-labelled hydroxymethylbilane synthase in its active form by multi-wavelength anomalous dispersion.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
5N29
 
 | |
5IZC
 
 | | Trypanosoma brucei PTR1 in complex with inhibitor F032 | | Descriptor: | ACETATE ION, GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Pozzi, C, Landi, G, Di Pisa, F, Mangani, S. | | Deposit date: | 2016-03-25 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Exploiting the 2-Amino-1,3,4-thiadiazole Scaffold To Inhibit Trypanosoma brucei Pteridine Reductase in Support of Early-Stage Drug Discovery.
ACS Omega, 2, 2017
|
|
4JB6
 
 | | Structure of Pseudomonas aeruginosa FabF mutant C164Q | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 2, POTASSIUM ION | | Authors: | Baum, B, Lecker, L. | | Deposit date: | 2013-02-19 | | Release date: | 2014-03-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of Pseudomonas aeruginosa beta-ketoacyl-(acyl-carrier-protein) synthase II (FabF) and a C164Q mutant provide templates for antibacterial drug discovery and identify a buried potassium ion and a ligand-binding site that is an artefact of the crystal form.
Acta Crystallogr F Struct Biol Commun, 71, 2015
|
|
4JPF
 
 | | Structure of wild type Pseudomonas aeruginosa FabF (KASII) in Complex with ligand | | Descriptor: | 3-(benzoylamino)-2-hydroxybenzoic acid, 3-oxoacyl-[acyl-carrier-protein] synthase 2, MAGNESIUM ION, ... | | Authors: | Baum, B, Brenk, R, Jaenicke, E. | | Deposit date: | 2013-03-19 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structures of Pseudomonas aeruginosa beta-ketoacyl-(acyl-carrier-protein) synthase II (FabF) and a C164Q mutant provide templates for antibacterial drug discovery and identify a buried potassium ion and a ligand-binding site that is an artefact of the crystal form.
Acta Crystallogr F Struct Biol Commun, 71, 2015
|
|
6TII
 
 | |
6TUD
 
 | |
1CVN
 
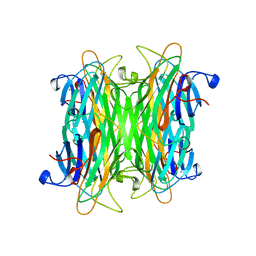 | | CONCANAVALIN A COMPLEXED TO TRIMANNOSIDE | | Descriptor: | CALCIUM ION, CONCANAVALIN A, MANGANESE (II) ION, ... | | Authors: | Naismith, J.H. | | Deposit date: | 1995-08-09 | | Release date: | 1996-10-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of trimannoside recognition by concanavalin A.
J.Biol.Chem., 271, 1996
|
|
4ETR
 
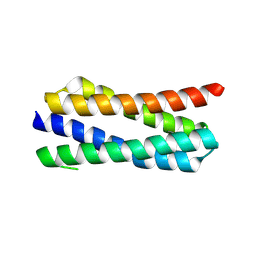 | | X-ray structure of PA2169 from Pseudomonas aeruginosa | | Descriptor: | Putative uncharacterized protein | | Authors: | Schnell, R, Sandalova, T, Lindqvist, Y, Schneider, G. | | Deposit date: | 2012-04-24 | | Release date: | 2013-01-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The AEROPATH project targeting Pseudomonas aeruginosa: crystallographic studies for assessment of potential targets in early-stage drug discovery.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
4ES6
 
 | |
4EXB
 
 | |
