2RAD
 
 | | Crystal structure of the succinoglycan biosynthesis protein. Northeast structural Genomics Consortium target BcR135 | | Descriptor: | Succinoglycan biosynthesis protein | | Authors: | Kuzin, A.P, Abashidze, M, Seetharaman, J, Wang, H, Mao, L, Cunningham, K, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-09-14 | | Release date: | 2007-10-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structure of the succinoglycan biosynthesis protein.
To be Published
|
|
4F2V
 
 | | Crystal Structure of de novo designed serine hydrolase, Northeast Structural Genomics Consortium (NESG) Target OR165 | | Descriptor: | DI(HYDROXYETHYL)ETHER, DODECYL-ALPHA-D-MALTOSIDE, De novo designed serine hydrolase | | Authors: | Kuzin, A, Lew, S, Seetharaman, J, Maglaqui, M, Xiao, R, Kohan, E, Rajagopalan, S, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-05-08 | | Release date: | 2012-05-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.493 Å) | | Cite: | Design of activated serine-containing catalytic triads with atomic-level accuracy.
Nat.Chem.Biol., 10, 2014
|
|
5GPL
 
 | | Crystal structure of Ccp1 | | Descriptor: | Putative nucleosome assembly protein C36B7.08c | | Authors: | Yin, F, Gao, F, Chen, Y. | | Deposit date: | 2016-08-03 | | Release date: | 2016-11-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Ccp1 Homodimer Mediates Chromatin Integrity by Antagonizing CENP-A Loading
Mol.Cell, 64, 2016
|
|
6D6Y
 
 | |
5GPK
 
 | | Crystal structure of Ccp1 mutant | | Descriptor: | Putative nucleosome assembly protein C36B7.08c | | Authors: | Yin, F, Gao, F, Chen, Y. | | Deposit date: | 2016-08-03 | | Release date: | 2016-11-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | Ccp1 Homodimer Mediates Chromatin Integrity by Antagonizing CENP-A Loading
Mol.Cell, 64, 2016
|
|
2HFD
 
 | | NMR structure of protein Hydrogenase-1 operon protein hyaE from Escherichia coli: Northeast Structural Genomics Consortium Target ER415 | | Descriptor: | Hydrogenase-1 operon protein hyaE | | Authors: | Singarapu, K.K, Liu, G, Eletsky, A, Parish, D, Atreya, H.S, Xu, D, Janjua, H, Cunningham, K, Ma, L.C, Xiao, R, Liu, J, Baran, M, Swapna, G.V.T, Acton, T, Rost, B, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-06-23 | | Release date: | 2006-08-22 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Protein chaperones Q8ZP25_SALTY from Salmonella typhimurium and HYAE_ECOLI from Escherichia coli exhibit thioredoxin-like structures despite lack of canonical thioredoxin active site sequence motif.
J.STRUCT.FUNCT.GENOM., 9, 2008
|
|
4ZKI
 
 | | The crystal structure of Histidine Kinase YycG with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Histidine kinase | | Authors: | Cai, Y. | | Deposit date: | 2015-04-30 | | Release date: | 2016-05-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.401 Å) | | Cite: | Conformational dynamics of the essential sensor histidine kinase WalK
Acta Crystallogr D Struct Biol, 73, 2017
|
|
8HF2
 
 | | Cryo-EM structure of WeiTsing | | Descriptor: | PRA1 family protein | | Authors: | Qin, L, Tang, L.H, Chen, Y.H. | | Deposit date: | 2022-11-09 | | Release date: | 2023-06-21 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4.14 Å) | | Cite: | WeiTsing, a pericycle-expressed ion channel, safeguards the stele to confer clubroot resistance.
Cell, 186, 2023
|
|
8CZJ
 
 | | A bacteria Zrt/Irt-like protein in the apo state | | Descriptor: | Putative membrane protein, SULFATE ION, [(Z)-octadec-9-enyl] (2R)-2,3-bis(oxidanyl)propanoate | | Authors: | Zhang, Y, Hu, J. | | Deposit date: | 2022-05-24 | | Release date: | 2023-02-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.751 Å) | | Cite: | Structural insights into the elevator-type transport mechanism of a bacterial ZIP metal transporter.
Nat Commun, 14, 2023
|
|
2BJY
 
 | |
2BKC
 
 | |
5C93
 
 | | Histidine kinase with ATP | | Descriptor: | Histidine kinase, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, SULFATE ION | | Authors: | Cai, Y. | | Deposit date: | 2015-06-26 | | Release date: | 2016-07-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.518 Å) | | Cite: | Conformational dynamics of the essential sensor histidine kinase WalK.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
8JCN
 
 | | The crystal structure of SARS-CoV-2 main protease in complex with Compound 58 | | Descriptor: | 1-[3-(diphenoxyphosphorylamino)phenyl]ethanone, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, ... | | Authors: | Zhao, Y, Zhu, Y, Rao, Z. | | Deposit date: | 2023-05-11 | | Release date: | 2024-05-15 | | Last modified: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | De novo design of SARS-CoV-2 main protease inhibitors with characteristic binding modes.
Structure, 32, 2024
|
|
8JCK
 
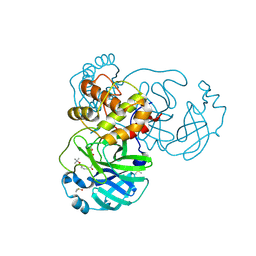 | | The crystal structure of SARS-CoV-2 main protease in complex with Compound 32 | | Descriptor: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Zhao, Y, Zhu, Y, Rao, Z. | | Deposit date: | 2023-05-11 | | Release date: | 2024-05-15 | | Last modified: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | De novo design of SARS-CoV-2 main protease inhibitors with characteristic binding modes.
Structure, 32, 2024
|
|
8JCM
 
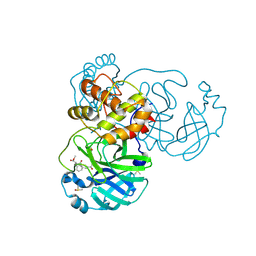 | | The crystal structure of SARS-CoV-2 main protease in complex with Compound 55 | | Descriptor: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, HYDROSULFURIC ACID, ... | | Authors: | Zhao, Y, Zhu, Y, Rao, Z. | | Deposit date: | 2023-05-11 | | Release date: | 2024-05-15 | | Last modified: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | De novo design of SARS-CoV-2 main protease inhibitors with characteristic binding modes.
Structure, 32, 2024
|
|
8JCO
 
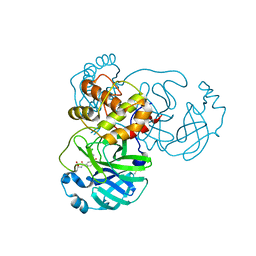 | |
8JCL
 
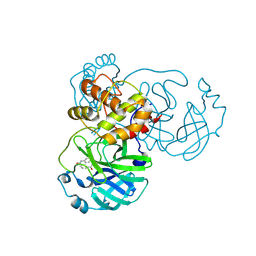 | |
8JCJ
 
 | | The crystal structure of SARS-CoV-2 main protease in complex with Compound 18 | | Descriptor: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Zhao, Y, Zhu, Y, Rao, Z. | | Deposit date: | 2023-05-11 | | Release date: | 2024-05-15 | | Last modified: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | De novo design of SARS-CoV-2 main protease inhibitors with characteristic binding modes.
Structure, 32, 2024
|
|
8ZY2
 
 | | Sarbecovirus BANAL-20-52 Spike Trimer in a Locked Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | Authors: | Wang, J, Xiong, X. | | Deposit date: | 2024-06-16 | | Release date: | 2025-02-05 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | SARS-related coronavirus S-protein structures reveal synergistic RBM interactions underpinning high-affinity human ACE2 binding.
Sci Adv, 11, 2025
|
|
8ZY9
 
 | | Ra9479 Bat ACE2 Dimer in Complex with Two BtKY72 Sarbecovirus Spike RBDs. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ... | | Authors: | Wang, J, Xiong, X. | | Deposit date: | 2024-06-16 | | Release date: | 2025-02-05 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | SARS-related coronavirus S-protein structures reveal synergistic RBM interactions underpinning high-affinity human ACE2 binding.
Sci Adv, 11, 2025
|
|
8ZY7
 
 | | Sarbecovirus HeB2013 Spike Trimer in a Locked Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Wang, J, Xiong, X. | | Deposit date: | 2024-06-16 | | Release date: | 2025-02-05 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | SARS-related coronavirus S-protein structures reveal synergistic RBM interactions underpinning high-affinity human ACE2 binding.
Sci Adv, 11, 2025
|
|
8ZY4
 
 | | Sarbecovirus YN2013 Spike Trimer in a Locked Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-2-[[5-[(Z)-(3-ethenyl-4-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1H-pyrrol-2-yl]methyl]-4-methyl-1H-pyrrol-3-yl]propanoic acid, ... | | Authors: | Wang, J, Xiong, X. | | Deposit date: | 2024-06-16 | | Release date: | 2025-02-05 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | SARS-related coronavirus S-protein structures reveal synergistic RBM interactions underpinning high-affinity human ACE2 binding.
Sci Adv, 11, 2025
|
|
8ZY6
 
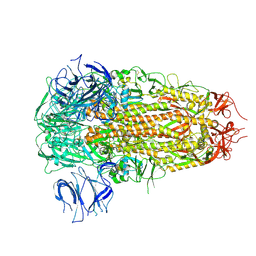 | | Sarbecovirus GX2013 Spike Trimer in a Locked Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Wang, J, Xiong, X. | | Deposit date: | 2024-06-16 | | Release date: | 2025-02-05 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | SARS-related coronavirus S-protein structures reveal synergistic RBM interactions underpinning high-affinity human ACE2 binding.
Sci Adv, 11, 2025
|
|
8ZY3
 
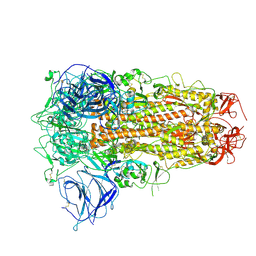 | | Sarbecovirus BANAL-20-236 Spike Trimer in a Locked Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Wang, J, Xiong, X. | | Deposit date: | 2024-06-16 | | Release date: | 2025-02-05 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | SARS-related coronavirus S-protein structures reveal synergistic RBM interactions underpinning high-affinity human ACE2 binding.
Sci Adv, 11, 2025
|
|
8ZY1
 
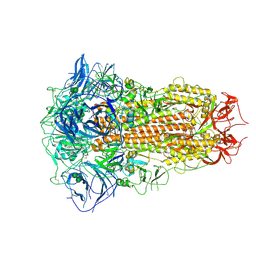 | | Sarbecovirus BM48-31 Spike Trimer in a Locked Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, ... | | Authors: | Wang, J, Xiong, X. | | Deposit date: | 2024-06-16 | | Release date: | 2025-02-05 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | SARS-related coronavirus S-protein structures reveal synergistic RBM interactions underpinning high-affinity human ACE2 binding.
Sci Adv, 11, 2025
|
|
