3TLE
 
 | |
2I8T
 
 | | GDP-mannose mannosyl hydrolase-calcium-GDP-mannose complex | | Descriptor: | CALCIUM ION, GDP-mannose mannosyl hydrolase, GLYCEROL, ... | | Authors: | Zou, Y, Li, C, Brunzelle, J.S, Nair, S.K. | | Deposit date: | 2006-09-03 | | Release date: | 2007-06-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Molecular basis for substrate selectivity and specificity by an LPS biosynthetic enzyme
Biochemistry, 46, 2007
|
|
2I8U
 
 | | GDP-mannose mannosyl hydrolase-calcium-GDP product complex | | Descriptor: | CALCIUM ION, GDP-mannose mannosyl hydrolase, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Zou, Y, Li, C, Brunzelle, J.S, Nair, S.K. | | Deposit date: | 2006-09-03 | | Release date: | 2007-06-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Molecular basis for substrate selectivity and specificity by an LPS biosynthetic enzyme
Biochemistry, 46, 2007
|
|
3M6I
 
 | | L-arabinitol 4-dehydrogenase | | Descriptor: | L-arabinitol 4-dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ZINC ION | | Authors: | Bae, B, Nair, S.K. | | Deposit date: | 2010-03-15 | | Release date: | 2010-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and engineering of L-arabinitol 4-dehydrogenase from Neurospora crassa
J.Mol.Biol., 402, 2010
|
|
3OEA
 
 | | Crystal structure of the Q121E mutants of C.polysaccharolyticus CBM16-1 bound to cellopentaose | | Descriptor: | CALCIUM ION, S-layer associated multidomain endoglucanase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Agarwal, V, Nair, S.K. | | Deposit date: | 2010-08-12 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Mutational insights into the roles of amino acid residues in ligand binding for two closely related family 16 carbohydrate binding modules.
J.Biol.Chem., 285, 2010
|
|
3IX4
 
 | | LasR-TP1 complex | | Descriptor: | 2,4-dibromo-6-({[(2-nitrophenyl)carbonyl]amino}methyl)phenyl 2-chlorobenzoate, Transcriptional activator protein lasR | | Authors: | Zou, Y, Nair, S.K. | | Deposit date: | 2009-09-03 | | Release date: | 2009-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular basis for the recognition of structurally distinct autoinducer mimics by the Pseudomonas aeruginosa LasR quorum-sensing signaling receptor.
Chem.Biol., 16, 2009
|
|
3IX8
 
 | | LasR-TP3 complex | | Descriptor: | 2,4-dibromo-6-({[(2-chlorophenyl)carbonyl]amino}methyl)phenyl 2-methylbenzoate, Transcriptional activator protein lasR | | Authors: | Zou, Y, Nair, S.K. | | Deposit date: | 2009-09-03 | | Release date: | 2009-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular basis for the recognition of structurally distinct autoinducer mimics by the Pseudomonas aeruginosa LasR quorum-sensing signaling receptor.
Chem.Biol., 16, 2009
|
|
3JPU
 
 | | LasR-TP4 complex | | Descriptor: | 4-bromo-2-({[(2-chlorophenyl)carbonyl]amino}methyl)-6-methylphenyl 2,4-dichlorobenzoate, Transcriptional activator protein lasR | | Authors: | Zou, Y, Nair, S.K. | | Deposit date: | 2009-09-04 | | Release date: | 2009-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular basis for the recognition of structurally distinct autoinducer mimics by the Pseudomonas aeruginosa LasR quorum-sensing signaling receptor.
Chem.Biol., 16, 2009
|
|
3OU2
 
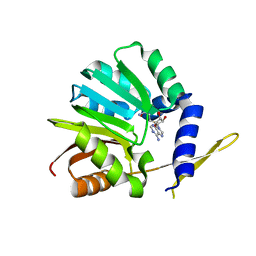 | | DhpI-SAH complex structure | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, SAM-dependent methyltransferase | | Authors: | Bae, B, Nair, S.K. | | Deposit date: | 2010-09-14 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Characterization and structure of DhpI, a phosphonate O-methyltransferase involved in dehydrophos biosynthesis.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3IX3
 
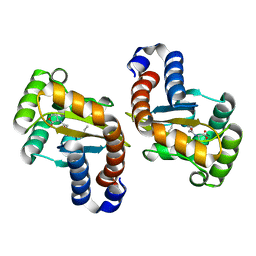 | | LasR-OC12 HSL complex | | Descriptor: | N-3-OXO-DODECANOYL-L-HOMOSERINE LACTONE, Transcriptional activator protein lasR | | Authors: | Zou, Y, Nair, S.K. | | Deposit date: | 2009-09-03 | | Release date: | 2009-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | LasR-OC12 HSL complex
TO BE PUBLISHED
|
|
3OEB
 
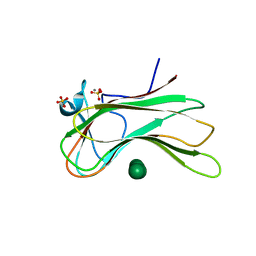 | |
3OO3
 
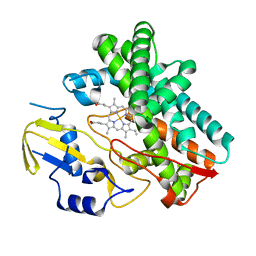 | |
3OU7
 
 | | DhpI-SAM-HEP complex | | Descriptor: | (2-hydroxyethyl)phosphonic acid, S-ADENOSYLMETHIONINE, SAM-dependent methyltransferase, ... | | Authors: | Bae, B, Nair, S.K. | | Deposit date: | 2010-09-14 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Characterization and structure of DhpI, a phosphonate O-methyltransferase involved in dehydrophos biosynthesis.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3OU6
 
 | | DhpI-SAM complex | | Descriptor: | S-ADENOSYLMETHIONINE, SAM-dependent methyltransferase, SULFATE ION | | Authors: | Bae, B, Nair, S.K. | | Deposit date: | 2010-09-14 | | Release date: | 2010-10-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Characterization and structure of DhpI, a phosphonate O-methyltransferase involved in dehydrophos biosynthesis.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3DFI
 
 | |
3DFK
 
 | |
3EUO
 
 | |
3CHI
 
 | |
3F8T
 
 | |
3CHU
 
 | | Crystal Structure of Di-iron Aurf | | Descriptor: | MU-OXO-DIIRON, p-Aminobenzoate N-Oxygenase | | Authors: | Zhang, H, Brunzelle, J.S, Nair, S.K. | | Deposit date: | 2008-03-10 | | Release date: | 2008-05-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | In vitro reconstitution and crystal structure of p-aminobenzoate N-oxygenase (AurF) involved in aureothin biosynthesis.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3CHT
 
 | |
3DFM
 
 | |
3EUQ
 
 | |
3EUT
 
 | |
3DFF
 
 | | The crystal structure of teicoplanin pseudoaglycone deacetylase Orf2 | | Descriptor: | GLYCEROL, TETRAETHYLENE GLYCOL, Teicoplanin pseudoaglycone deacetylases Orf2, ... | | Authors: | Zou, Y, Brunzelle, J.S, Nair, S.K. | | Deposit date: | 2008-06-11 | | Release date: | 2008-07-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of lipoglycopeptide antibiotic deacetylases: implications for the biosynthesis of a40926 and teicoplanin.
Chem.Biol., 15, 2008
|
|
