2DQW
 
 | |
2DUC
 
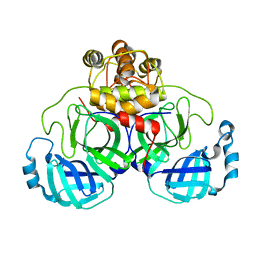 | | Crystal structure of SARS coronavirus main proteinase(3CLPRO) | | Descriptor: | Replicase polyprotein 1ab | | Authors: | Wang, H, Kim, Y.T, Muramatsu, T, Takemoto, C, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-07-21 | | Release date: | 2007-07-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | SARS-CoV 3CL protease cleaves its C-terminal autoprocessing site by novel subsite cooperativity
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
2DVL
 
 | |
2DWB
 
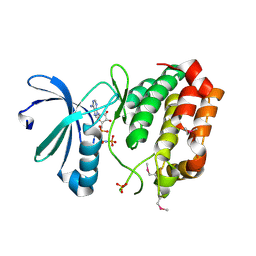 | | Aurora-A kinase complexed with AMPPNP | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, SULFATE ION, Serine/threonine-protein kinase 6 | | Authors: | Kukimoto-Niino, M, Murayama, K, Shirouzu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-08-10 | | Release date: | 2007-07-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Aurora-A kinase complexed with AMPPNP
To be Published
|
|
2DQ0
 
 | | Crystal structure of seryl-tRNA synthetase from Pyrococcus horikoshii complexed with a seryl-adenylate analog | | Descriptor: | 5'-O-(N-(L-SERYL)-SULFAMOYL)ADENOSINE, SULFATE ION, Seryl-tRNA synthetase | | Authors: | Itoh, Y, Sekine, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-05-18 | | Release date: | 2007-06-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystallographic and mutational studies of seryl-tRNA synthetase from the archaeon Pyrococcus horikoshii.
Rna Biol., 5, 2008
|
|
2DJ6
 
 | |
2E18
 
 | |
2E3V
 
 | | Crystal structure of the first fibronectin type III domain of neural cell adhesion molecule splicing isoform from human muscle culture lambda-4.4 | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Nishino, A, Saijo, S, Kishishita, S, Chen, L, Liu, Z.J, Wang, B.C, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-11-30 | | Release date: | 2007-06-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of the first fibronectin type III domain of neural cell adhesion molecule splicing isoform from human muscle culture lambda-4.4
To be Published
|
|
2E8B
 
 | | Crystal structure of the putative protein (Aq1419) from Aquifex aeolicus VF5 | | Descriptor: | Probable molybdopterin-guanine dinucleotide biosynthesis protein A | | Authors: | Kumarevel, T.S, Malathy sony, S.M, Ponnuswamy, M.N, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-01-19 | | Release date: | 2008-01-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Crystal structure of the putative protein (Aq1419) from Aquifex aeolicus VF5
To be Published
|
|
2E9Y
 
 | |
2EB0
 
 | |
2ENW
 
 | |
2EQ5
 
 | |
2DXA
 
 | | Crystal structure of trans editing enzyme ProX from E.coli | | Descriptor: | CHLORIDE ION, Protein ybaK | | Authors: | Kato-Murayama, M, Murayama, K, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-08-25 | | Release date: | 2007-02-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal structure of trans editing enzyme ProX from E.coli
To be Published
|
|
2DYC
 
 | | Crystal structure of the N-terminal domain of mouse galectin-4 | | Descriptor: | Galectin-4 | | Authors: | Kato-Murayama, M, Murayama, K, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-09-08 | | Release date: | 2007-10-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the N-terminal domain of mouse galectin-4
To be Published
|
|
2E1H
 
 | |
2EAY
 
 | |
2EGZ
 
 | | Crystal structure of the 3-dehydroquinate dehydratase from Aquifex aeolicus VF5 | | Descriptor: | 3-dehydroquinate dehydratase, L(+)-TARTARIC ACID | | Authors: | Karthe, P, Kumarevel, T.S, Ebihara, A, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-02 | | Release date: | 2007-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of the 3-dehydroquinate dehydratase from Aquifex aeolicus VF5
To be Published
|
|
2EHU
 
 | | Crystal analysis of 1-pyrroline-5-carboxylate dehydrogenase from thermus with bound NAD and Inhibitor L-serine | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1-pyrroline-5-carboxylate dehydrogenase, ACETATE ION, ... | | Authors: | Inagaki, E, Sakamoto, K, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-09 | | Release date: | 2007-09-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure Analysis of delta1-pyrroline-5-carboxylate dehydrogenase in ternary complex with inhibitor and NAD.
To be Published
|
|
2EJ6
 
 | | Crystal analysis of delta1-pyrroline-5-carboxylate dehydrogenase from Thermus thermophilus with bound D-proline | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1-pyrroline-5-carboxylate dehydrogenase, ACETATE ION, ... | | Authors: | Inagaki, E, Sakamoto, K, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-15 | | Release date: | 2007-09-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal structure analysis of delta1-pyrroline-5-carboxylate dehydrogenase in ternary complex with inhibitor and NAD
To be Published
|
|
2DXF
 
 | | Crystal structure of nucleoside diphosphate kinase in complex with GTP analog | | Descriptor: | CHLORIDE ION, Nucleoside diphosphate kinase, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Kato-Murayama, M, Murayama, K, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-08-25 | | Release date: | 2007-02-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of nucleoside diphosphate kinase in complex with GTP analog
To be Published
|
|
2DY9
 
 | | Crystal structure of nucleoside diphosphate kinase in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Kato-Murayama, M, Murayama, K, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-09-08 | | Release date: | 2007-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structure of nucleoside diphosphate kinase in complex with ADP
To be Published
|
|
2DYK
 
 | | Crystal structure of N-terminal GTP-binding domain of EngA from Thermus thermophilus HB8 | | Descriptor: | GTP-binding protein, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Kawazoe, M, Takemoto, C, Fusatomi, E, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-09-14 | | Release date: | 2007-03-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal structure of N-terminal GTP-binding domain of EngA from Thermus thermophilus HB8
to be published
|
|
2DYL
 
 | | Crystal structure of human mitogen-activated protein kinase kinase 7 activated mutant (S287D, T291D) | | Descriptor: | Dual specificity mitogen-activated protein kinase kinase 7 | | Authors: | Kukimoto-Niino, M, Takagi, T, Kaminishi, T, Uchikubo-Kamo, T, Terada, T, Matsuzaki, O, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-09-15 | | Release date: | 2007-08-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of human mitogen-activated protein kinase kinase 7 activated mutant (S287D, T291D)
To be Published
|
|
2DYQ
 
 | | Crystal Structure of the C-terminal Phophotyrosine Interaction Domain of Human APBB3 | | Descriptor: | Amyloid beta A4 precursor protein-binding family B member 3 | | Authors: | Nishino, A, Saijo, S, Kishishita, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-09-16 | | Release date: | 2007-09-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal Structure of the C-terminal Phophotyrosine Interaction Domain of Human APBB3
To be Published
|
|
