8CW7
 
 | | 200us Temperature-Jump (Dark2) XFEL structure of Lysozyme | | Descriptor: | ACETATE ION, CHLORIDE ION, Lysozyme C, ... | | Authors: | Wolff, A.M, Thompson, M.C, Fraser, J.S, Nango, E. | | Deposit date: | 2022-05-18 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Mapping protein dynamics at high spatial resolution with temperature-jump X-ray crystallography.
Nat.Chem., 15, 2023
|
|
8CVU
 
 | | 20ns Temperature-Jump (Light) XFEL structure of Lysozyme | | Descriptor: | ACETATE ION, CHLORIDE ION, Lysozyme C, ... | | Authors: | Wolff, A.M, Thompson, M.C, Fraser, J.S, Nango, E. | | Deposit date: | 2022-05-18 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Mapping protein dynamics at high spatial resolution with temperature-jump X-ray crystallography.
Nat.Chem., 15, 2023
|
|
8CW6
 
 | | 200us Temperature-Jump (Dark1) XFEL structure of Lysozyme | | Descriptor: | ACETATE ION, CHLORIDE ION, Lysozyme C, ... | | Authors: | Wolff, A.M, Thompson, M.C, Fraser, J.S, Nango, E. | | Deposit date: | 2022-05-18 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Mapping protein dynamics at high spatial resolution with temperature-jump X-ray crystallography.
Nat.Chem., 15, 2023
|
|
8CW8
 
 | | Laser Off Temperature-Jump XFEL structure of Lysozyme | | Descriptor: | ACETATE ION, CHLORIDE ION, Lysozyme C, ... | | Authors: | Wolff, A.M, Thompson, M.C, Fraser, J.S, Nango, E. | | Deposit date: | 2022-05-18 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Mapping protein dynamics at high spatial resolution with temperature-jump X-ray crystallography.
Nat.Chem., 15, 2023
|
|
8CW1
 
 | | 20us Temperature-Jump (Dark1) XFEL structure of Lysozyme | | Descriptor: | ACETATE ION, CHLORIDE ION, Lysozyme C, ... | | Authors: | Wolff, A.M, Thompson, M.C, Fraser, J.S, Nango, E. | | Deposit date: | 2022-05-18 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Mapping protein dynamics at high spatial resolution with temperature-jump X-ray crystallography.
Nat.Chem., 15, 2023
|
|
8CW3
 
 | | 20us Temperature-Jump (Dark2) XFEL structure of Lysozyme | | Descriptor: | ACETATE ION, CHLORIDE ION, Lysozyme C, ... | | Authors: | Wolff, A.M, Thompson, M.C, Fraser, J.S, Nango, E. | | Deposit date: | 2022-05-18 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Mapping protein dynamics at high spatial resolution with temperature-jump X-ray crystallography.
Nat.Chem., 15, 2023
|
|
8CWG
 
 | | 200us Temperature-Jump (Dark1) XFEL structure of Lysozyme Bound to N,N'-diacetylchitobiose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, CHLORIDE ION, Lysozyme C, ... | | Authors: | Wolff, A.M, Thompson, M.C, Fraser, J.S, Nango, E. | | Deposit date: | 2022-05-19 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mapping protein dynamics at high spatial resolution with temperature-jump X-ray crystallography.
Nat.Chem., 15, 2023
|
|
8CWB
 
 | | Laser Off Temperature-Jump XFEL structure of Lysozyme Bound to N,N'-diacetylchitobiose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, CHLORIDE ION, Lysozyme C, ... | | Authors: | Wolff, A.M, Thompson, M.C, Fraser, J.S, Nango, E. | | Deposit date: | 2022-05-19 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Mapping protein dynamics at high spatial resolution with temperature-jump X-ray crystallography.
Nat.Chem., 15, 2023
|
|
8CWE
 
 | | 20ns Temperature-Jump (Dark2) XFEL structure of Lysozyme Bound to N,N'-diacetylchitobiose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, CHLORIDE ION, Lysozyme C, ... | | Authors: | Wolff, A.M, Thompson, M.C, Fraser, J.S, Nango, E. | | Deposit date: | 2022-05-19 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Mapping protein dynamics at high spatial resolution with temperature-jump X-ray crystallography.
Nat.Chem., 15, 2023
|
|
8CVV
 
 | | 20ns Temperature-Jump (Dark1) XFEL structure of Lysozyme | | Descriptor: | ACETATE ION, CHLORIDE ION, Lysozyme C, ... | | Authors: | Wolff, A.M, Thompson, M.C, Fraser, J.S, Nango, E. | | Deposit date: | 2022-05-18 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Mapping protein dynamics at high spatial resolution with temperature-jump X-ray crystallography.
Nat.Chem., 15, 2023
|
|
8CWD
 
 | | 20ns Temperature-Jump (Dark1) XFEL structure of Lysozyme Bound to N,N'-diacetylchitobiose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, CHLORIDE ION, Lysozyme C, ... | | Authors: | Wolff, A.M, Thompson, M.C, Fraser, J.S, Nango, E. | | Deposit date: | 2022-05-19 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Mapping protein dynamics at high spatial resolution with temperature-jump X-ray crystallography.
Nat.Chem., 15, 2023
|
|
8CVW
 
 | | 20ns Temperature-Jump (Dark2) XFEL structure of Lysozyme | | Descriptor: | ACETATE ION, CHLORIDE ION, Lysozyme C, ... | | Authors: | Wolff, A.M, Thompson, M.C, Fraser, J.S, Nango, E. | | Deposit date: | 2022-05-18 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Mapping protein dynamics at high spatial resolution with temperature-jump X-ray crystallography.
Nat.Chem., 15, 2023
|
|
8CWF
 
 | | 200us Temperature-Jump (Light) XFEL structure of Lysozyme Bound to N,N'-diacetylchitobiose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, CHLORIDE ION, Lysozyme C, ... | | Authors: | Wolff, A.M, Thompson, M.C, Fraser, J.S, Nango, E. | | Deposit date: | 2022-05-19 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Mapping protein dynamics at high spatial resolution with temperature-jump X-ray crystallography.
Nat.Chem., 15, 2023
|
|
8CW5
 
 | | 200us Temperature-Jump (Light) XFEL structure of Lysozyme | | Descriptor: | ACETATE ION, CHLORIDE ION, Lysozyme C, ... | | Authors: | Wolff, A.M, Thompson, M.C, Fraser, J.S, Nango, E. | | Deposit date: | 2022-05-18 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Mapping protein dynamics at high spatial resolution with temperature-jump X-ray crystallography.
Nat.Chem., 15, 2023
|
|
8CWC
 
 | | 20ns Temperature-Jump (Light) XFEL structure of Lysozyme Bound to N,N'-diacetylchitobiose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, CHLORIDE ION, Lysozyme C, ... | | Authors: | Wolff, A.M, Thompson, M.C, Fraser, J.S, Nango, E. | | Deposit date: | 2022-05-19 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Mapping protein dynamics at high spatial resolution with temperature-jump X-ray crystallography.
Nat.Chem., 15, 2023
|
|
8FG7
 
 | |
8FRA
 
 | | Mouse acidic mammalian chitinase, catalytic domain in complex with diacetylchitobiose at pH 5.60 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Acidic mammalian chitinase, ... | | Authors: | Diaz, R.E, Fraser, J.S. | | Deposit date: | 2023-01-06 | | Release date: | 2023-03-08 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural characterization of ligand binding and pH-specific enzymatic activity of mouse Acidic Mammalian Chitinase.
Biorxiv, 2024
|
|
8FRC
 
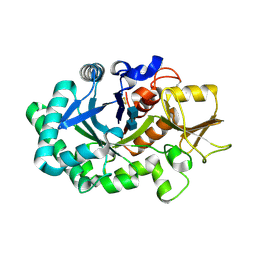 | |
8FRB
 
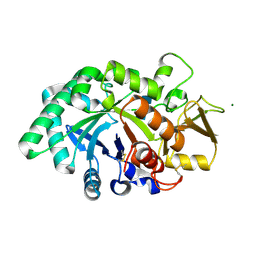 | | Mouse acidic mammalian chitinase, catalytic domain in complex with N,N'-diacetylchitobiose at pH 5.25 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetylamino-2-deoxy-alpha-L-idopyranose, ... | | Authors: | Diaz, R.E, Fraser, J.S. | | Deposit date: | 2023-01-06 | | Release date: | 2023-03-08 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural characterization of ligand binding and pH-specific enzymatic activity of mouse Acidic Mammalian Chitinase.
Biorxiv, 2024
|
|
8FRD
 
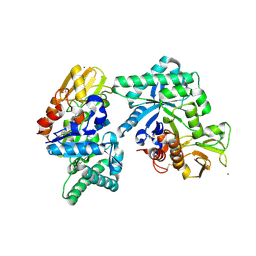 | |
5S18
 
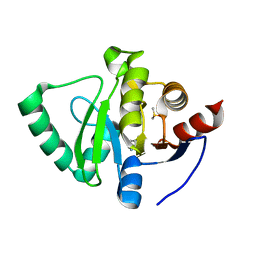 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with EN300-321461 | | Descriptor: | 6,7-dihydro-5H-pyrrolo[2,3-d]pyrimidine, DIMETHYL SULFOXIDE, Non-structural protein 3 | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5S20
 
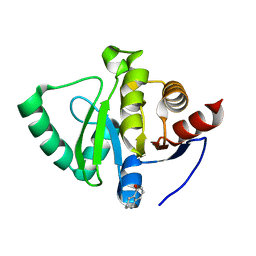 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with PB1827975385 | | Descriptor: | (5R)-5-amino-5,6,7,8-tetrahydronaphthalen-1-ol, Non-structural protein 3 | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.037 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5S2K
 
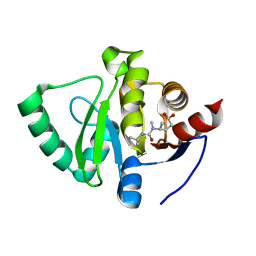 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with Z445856640 | | Descriptor: | N-[(3R)-1,1-dioxo-1lambda~6~-thiolan-3-yl]-N-methyl-N'-propan-2-ylurea, Non-structural protein 3 | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.097 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5S30
 
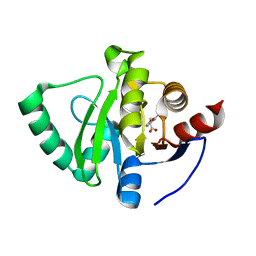 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with Z65532537 | | Descriptor: | (2R)-2-(2-fluorophenoxy)propanoic acid, Non-structural protein 3 | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5S3K
 
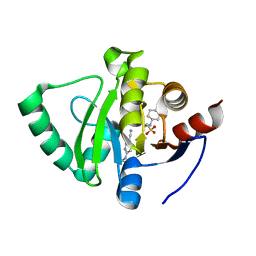 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with Z219104216 | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, 6-(ethylamino)pyridine-3-carbonitrile, Non-structural protein 3 | | Authors: | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | Deposit date: | 2020-11-02 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
