1V49
 
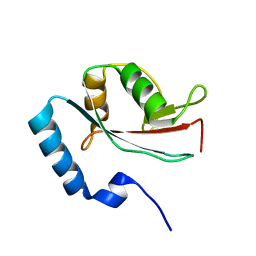 | | Solution structure of microtubule-associated protein light chain-3 | | Descriptor: | Microtubule-associated proteins 1A/1B light chain 3B | | Authors: | Kouno, T, Mizuguchi, M, Tanida, I, Ueno, T, Kominami, E, Kawano, K. | | Deposit date: | 2003-11-11 | | Release date: | 2004-12-28 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of microtubule-associated protein light chain 3 and identification of its functional subdomains.
J.Biol.Chem., 280, 2005
|
|
2YKI
 
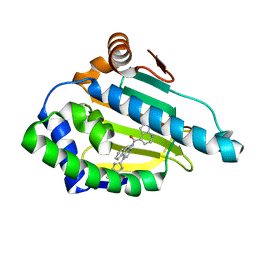 | | Tricyclic series of Hsp90 inhibitors | | Descriptor: | 1-H-PYRROLO[2,3-B]PYRIDINE-4-CARBOXYLIC ACID [4-(3H-IMIDAZO[4,5-C]PYRIDIN-2-YL)-9H-FLUOREN-9-YL]-AMIDE, HEAT SHOCK PROTEIN HSP 90-ALPHA | | Authors: | Dupuy, A, Vallee, F. | | Deposit date: | 2011-05-27 | | Release date: | 2011-10-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Tricyclic Series of Heat Shock Protein 90 (Hsp90) Inhibitors Part I: Discovery of Tricyclic Imidazo[4,5-C]Pyridines as Potent Inhibitors of the Hsp90 Molecular Chaperone.
J.Med.Chem., 54, 2011
|
|
2YK9
 
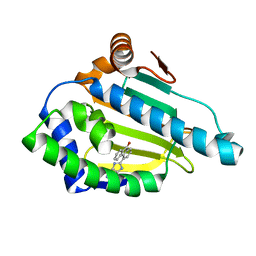 | | Tricyclic series of Hsp90 inhibitors | | Descriptor: | 4-(1H-IMIDAZO[4,5-C]PYRIDIN-2-YL)FLUOREN-9-ONE, HEAT SHOCK PROTEIN HSP 90-ALPHA | | Authors: | Dupuy, A, Vallee, F. | | Deposit date: | 2011-05-26 | | Release date: | 2011-10-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Tricyclic Series of Heat Shock Protein 90 (Hsp90) Inhibitors Part I: Discovery of Tricyclic Imidazo[4,5-C]Pyridines as Potent Inhibitors of the Hsp90 Molecular Chaperone.
J.Med.Chem., 54, 2011
|
|
2YKB
 
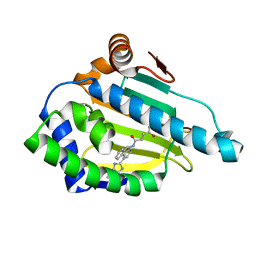 | | Tricyclic series of Hsp90 inhibitors | | Descriptor: | HEAT SHOCK PROTEIN HSP 90-ALPHA, N-[4-(3H-IMIDAZO[4,5-C]PYRIDIN-2-YL)-9H-FLUOREN-9-YL]-SUCCINAMIDE | | Authors: | Dupuy, A, Vallee, F. | | Deposit date: | 2011-05-26 | | Release date: | 2011-10-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Tricyclic Series of Heat Shock Protein 90 (Hsp90) Inhibitors Part I: Discovery of Tricyclic Imidazo[4,5-C]Pyridines as Potent Inhibitors of the Hsp90 Molecular Chaperone.
J.Med.Chem., 54, 2011
|
|
2YKC
 
 | | Tricyclic series of Hsp90 inhibitors | | Descriptor: | HEAT SHOCK PROTEIN HSP 90-ALPHA, N-[4-(3H-IMIDAZO[4,5-C]PYRIDIN-2-YL)-9H-FLUOREN-9-YL-ISONICOTINAMIDE | | Authors: | Dupuy, A, Vallee, F. | | Deposit date: | 2011-05-26 | | Release date: | 2011-10-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Tricyclic Series of Heat Shock Protein 90 (Hsp90) Inhibitors Part I: Discovery of Tricyclic Imidazo[4,5-C]Pyridines as Potent Inhibitors of the Hsp90 Molecular Chaperone.
J.Med.Chem., 54, 2011
|
|
2D4R
 
 | | Crystal structure of TTHA0849 from Thermus thermophilus HB8 | | Descriptor: | SULFATE ION, hypothetical protein TTHA0849 | | Authors: | Nakabayashi, M, Shibata, N, Kuramitsu, S, Higuchi, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-10-23 | | Release date: | 2005-12-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of a conserved hypothetical protein, TTHA0849 from Thermus thermophilus HB8, at 2.4 A resolution: a putative member of the StAR-related lipid-transfer (START) domain superfamily.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
2E67
 
 | |
2E2F
 
 | |
2YXG
 
 | |
2YZT
 
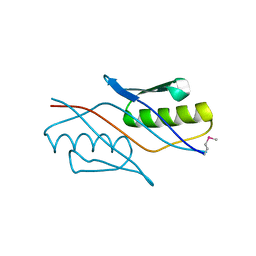 | |
3W5N
 
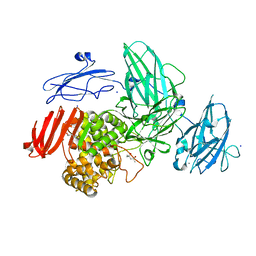 | | Crystal Structure of Streptomyces avermitilis alpha-L-rhamnosidase complexed with L-rhamnose | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, Putative rhamnosidase, ... | | Authors: | Fujimoto, Z, Jackson, A, Michikawa, M, Maehara, T, Momma, M, Henrissat, B.F, Gilbert, H.J, Kaneko, S. | | Deposit date: | 2013-01-31 | | Release date: | 2013-03-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of a Streptomyces avermitilis alpha-L-rhamnosidase reveals a novel carbohydrate-binding module CBM67 within the six-domain arrangement.
J.Biol.Chem., 288, 2013
|
|
3W5M
 
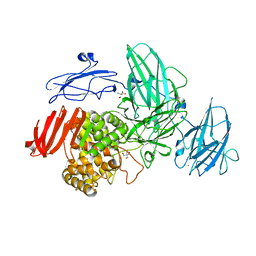 | | Crystal Structure of Streptomyces avermitilis alpha-L-rhamnosidase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, Putative rhamnosidase | | Authors: | Fujimoto, Z, Jackson, A, Michikawa, M, Maehara, T, Momma, M, Henrissat, B.F, Gilbert, H.J, Kaneko, S. | | Deposit date: | 2013-01-31 | | Release date: | 2013-03-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of a Streptomyces avermitilis alpha-L-rhamnosidase reveals a novel carbohydrate-binding module CBM67 within the six-domain arrangement.
J.Biol.Chem., 288, 2013
|
|
2D56
 
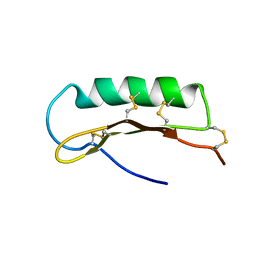 | | Solution Structure of ASABF, Antibacterial Peptide Isolated from a Nematode, Ascaris Suum | | Descriptor: | ASABF | | Authors: | Nakano, M, Aizawa, T, Kamiya, M, Miura, K, Kumaki, Y, Demura, M, Tsuda, S, Kawano, K. | | Deposit date: | 2005-10-28 | | Release date: | 2006-11-14 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution structure and function of ASABFd18c, antibacterial peptide isolated from a nematode, asucaris suum
To be Published
|
|
