8PNW
 
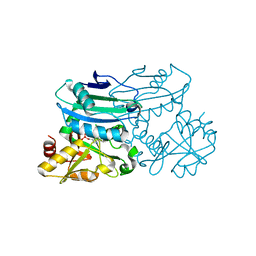 | | Crystal structure of D-amino acid aminotransferase from Blastococcus saxobsidens in holo form with PLP | | 分子名称: | Branched-chain amino acid aminotransferase/4-amino-4-deoxychorismate lyase, CHLORIDE ION, PYRIDOXAL-5'-PHOSPHATE | | 著者 | Matyuta, I.O, Boyko, K.M, Nikolaeva, A.Y, Shilova, S.A, Popov, V.O. | | 登録日 | 2023-07-03 | | 公開日 | 2023-10-25 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Expanded Substrate Specificity in D-Amino Acid Transaminases: A Case Study of Transaminase from Blastococcus saxobsidens.
Int J Mol Sci, 24, 2023
|
|
8PNY
 
 | | Crystal structure of D-amino acid aminotransferase from Blastococcus saxobsidens complexed with phenylhydrazine and in its apo form | | 分子名称: | Branched-chain amino acid aminotransferase/4-amino-4-deoxychorismate lyase, [6-methyl-5-oxidanyl-4-[(2-phenylhydrazinyl)methyl]pyridin-3-yl]methyl dihydrogen phosphate | | 著者 | Matyuta, I.O, Boyko, K.M, Nikolaeva, A.Y, Shilova, S.A, Popov, V.O. | | 登録日 | 2023-07-03 | | 公開日 | 2023-10-25 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Expanded Substrate Specificity in D-Amino Acid Transaminases: A Case Study of Transaminase from Blastococcus saxobsidens.
Int J Mol Sci, 24, 2023
|
|
8PNX
 
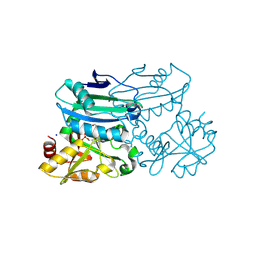 | | Crystal structure of D-amino acid aminotransferase from Blastococcus saxobsidens in PMP form | | 分子名称: | 1,2-ETHANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Branched-chain amino acid aminotransferase/4-amino-4-deoxychorismate lyase, ... | | 著者 | Matyuta, I.O, Boyko, K.M, Nikolaeva, A.Y, Shilova, S.A, Popov, V.O. | | 登録日 | 2023-07-03 | | 公開日 | 2024-07-10 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural determinants of dual substrate recognition in the transaminase from Blastococcus saxobsidens specific to D-amino acids and R-amines
To Be Published
|
|
8ONN
 
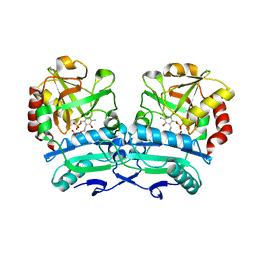 | | Crystal structure of D-amino acid aminotransferase from Aminobacterium colombiense point mutant E113A complexed with 3-aminooxypropionic acid | | 分子名称: | 3-[(~{E})-[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylideneamino]oxypropanoic acid, Aminotransferase class IV | | 著者 | Matyuta, I.O, Boyko, K.M, Minyaev, M.E, Shilova, S.A, Bezsudnova, E.Y, Popov, V.O. | | 登録日 | 2023-04-03 | | 公開日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | In search for structural targets for engineering d-amino acid transaminase: modulation of pH optimum and substrate specificity.
Biochem.J., 480, 2023
|
|
8ONL
 
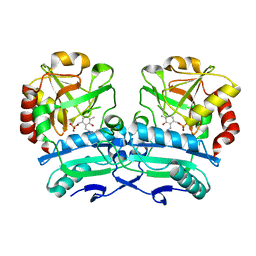 | | Crystal structure of D-amino acid aminotransferase from Aminobacterium colombiense point mutant E113A | | 分子名称: | Aminotransferase class IV, PYRIDOXAL-5'-PHOSPHATE | | 著者 | Matyuta, I.O, Boyko, K.M, Minyaev, M.E, Shilova, S.A, Bezsudnova, E.Y, Popov, V.O. | | 登録日 | 2023-04-03 | | 公開日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | In search for structural targets for engineering d-amino acid transaminase: modulation of pH optimum and substrate specificity.
Biochem.J., 480, 2023
|
|
8ONJ
 
 | | Crystal structure of D-amino acid aminotransferase from Aminobacterium colombiense point mutant R88L | | 分子名称: | Aminotransferase class IV, DI(HYDROXYETHYL)ETHER, PYRIDOXAL-5'-PHOSPHATE | | 著者 | Matyuta, I.O, Boyko, K.M, Minyaev, M.E, Shilova, S.A, Bezsudnova, E.Y, Popov, V.O. | | 登録日 | 2023-04-03 | | 公開日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | In search for structural targets for engineering d-amino acid transaminase: modulation of pH optimum and substrate specificity.
Biochem.J., 480, 2023
|
|
8ONM
 
 | | Crystal structure of D-amino acid aminotransferase from Aminobacterium colombiense point mutant E113A complexed with D-glutamate | | 分子名称: | (~{Z})-2-[[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylamino]pent-2-enedioic acid, 1,2-ETHANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, ... | | 著者 | Matyuta, I.O, Boyko, K.M, Minyaev, M.E, Shilova, S.A, Bezsudnova, E.Y, Popov, V.O. | | 登録日 | 2023-04-03 | | 公開日 | 2024-04-10 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Probing of the structural and catalytic roles of the residues in the active site of transaminase from Aminobacterium colombiense
To Be Published
|
|
8AIE
 
 | | Crystal structure of D-amino acid aminotransferase from Aminobacterium colombiense complexed with D-cycloserine | | 分子名称: | 3-azanyloxy-2-[(~{E})-[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylideneamino]propanoic acid, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Aminotransferase class IV, ... | | 著者 | Matyuta, I.O, Boyko, K.M, Nikolaeva, A.Y, Shilova, S.A, Popov, V.O, Bezsudnova, E.Y. | | 登録日 | 2022-07-26 | | 公開日 | 2022-11-16 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | 3D Structure of D-Аmino Acid Тransaminase from Aminobacterium colombiense in Complex with D-Cycloserine
Crystallography Reports, 68, 2023
|
|
8AHR
 
 | | Crystal structure of D-amino acid aminotransferase from Aminobacterium colombiense in holo form with PLP | | 分子名称: | Aminotransferase class IV, PYRIDOXAL-5'-PHOSPHATE | | 著者 | Matyuta, I.O, Boyko, K.M, Nikolaeva, A.Y, Shilova, S.A, Rakitina, T.V, Popov, V.O, Bezsudnova, E.Y. | | 登録日 | 2022-07-22 | | 公開日 | 2022-08-03 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | To the Understanding of Catalysis by D-Amino Acid Transaminases: A Case Study of the Enzyme from Aminobacterium colombiense.
Molecules, 28, 2023
|
|
8AYK
 
 | | Crystal structure of D-amino acid aminotrensferase from Aminobacterium colombiense complexed with D-glutamate | | 分子名称: | (~{Z})-2-[[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylamino]pent-2-enedioic acid, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Aminotransferase class IV | | 著者 | Matyuta, I.O, Boyko, K.M, Nikolaeva, A.Y, Shilova, S.A, Rakitina, T.V, Minyaev, M.E, Popov, V.O, Bezsudnova, E.Y. | | 登録日 | 2022-09-02 | | 公開日 | 2022-11-16 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | To the Understanding of Catalysis by D-Amino Acid Transaminases: A Case Study of the Enzyme from Aminobacterium colombiense.
Molecules, 28, 2023
|
|
8AYJ
 
 | | Crystal structure of D-amino acid aminotransferase from Aminobacterium colombiens complexed with 3-aminooxypropionic acid | | 分子名称: | 1,2-ETHANEDIOL, 3-[(~{E})-[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylideneamino]oxypropanoic acid, Aminotransferase class IV, ... | | 著者 | Matyuta, I.O, Boyko, K.M, Nikolaeva, A.Y, Shilova, S.A, Rakitina, T.V, Popov, V.O, Bezsudnova, E.Y. | | 登録日 | 2022-09-02 | | 公開日 | 2022-11-16 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | In search for structural targets for engineering d-amino acid transaminase: modulation of pH optimum and substrate specificity.
Biochem.J., 480, 2023
|
|
