5WQW
 
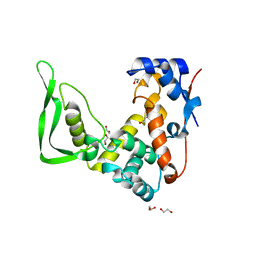 | | X-ray structure of catalytic domain of autolysin from Clostridium perfringens | | 分子名称: | 1,2-ETHANEDIOL, N-acetylglucosaminidase | | 著者 | Tamai, E, Sekiya, H, Goda, E, Makihata, N, Maki, J, Yoshida, H, Kamitori, S. | | 登録日 | 2016-11-29 | | 公開日 | 2016-12-07 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | Structural and biochemical characterization of the Clostridium perfringens autolysin catalytic domain
FEBS Lett., 591, 2017
|
|
8YXK
 
 | |
8YXN
 
 | |
7F5I
 
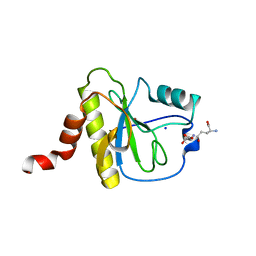 | | X-ray structure of Clostridium perfringens-specific amidase endolysin | | 分子名称: | GLUTAMIC ACID, SODIUM ION, ZINC ION, ... | | 著者 | Kamitori, S, Tamai, E. | | 登録日 | 2021-06-22 | | 公開日 | 2022-05-04 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Structural and biochemical characterization of the Clostridium perfringens-specific Zn 2+ -dependent amidase endolysin, Psa, catalytic domain.
Biochem.Biophys.Res.Commun., 576, 2021
|
|
7CFL
 
 | |
4KRT
 
 | |
4KRU
 
 | |
5B23
 
 | |
8GSY
 
 | |
8GSX
 
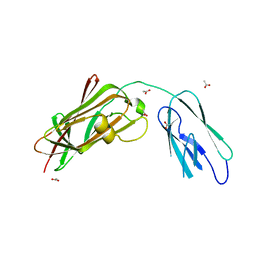 | |
5YFK
 
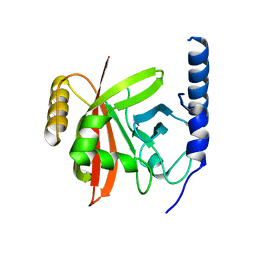 | |
6IXZ
 
 | |
6IXY
 
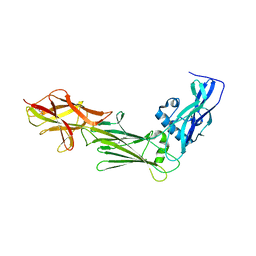 | |
5XCC
 
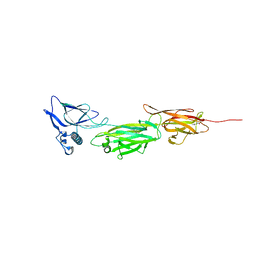 | |
5XCB
 
 | |
7D6P
 
 | |
7D6T
 
 | |
