2JMY
 
 | | Solution structure of CM15 in DPC micelles | | 分子名称: | CM15 | | 著者 | Respondek, M, Madl, T, Goebl, C, Golser, R, Zangger, K. | | 登録日 | 2006-12-13 | | 公開日 | 2007-07-17 | | 最終更新日 | 2024-05-08 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Mapping the orientation of helices in micelle-bound peptides by paramagnetic relaxation waves
J.Am.Chem.Soc., 129, 2007
|
|
2H3C
 
 | | Structural basis for nucleic acid and toxin recognition of the bacterial antitoxin CcdA | | 分子名称: | 5'-D(P*AP*TP*AP*TP*GP*TP*AP*TP*AP*CP*CP*CP*G)-3', 5'-D(P*TP*CP*GP*GP*GP*TP*AP*TP*AP*CP*AP*TP*A)-3', CcdA | | 著者 | Madl, T, Van Melderen, L, Respondek, M, Oberer, M, Keller, W, Zangger, K. | | 登録日 | 2006-05-22 | | 公開日 | 2006-11-21 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural Basis for Nucleic Acid and Toxin Recognition of the Bacterial Antitoxin CcdA
J.Mol.Biol., 364, 2006
|
|
2H3A
 
 | | Structural basis for nucleic acid and toxin recognition of the bacterial antitoxin CcdA | | 分子名称: | 5'-D(P*AP*TP*AP*TP*GP*TP*AP*TP*AP*CP*CP*CP*G)-3', 5'-D(P*TP*CP*GP*GP*GP*TP*AP*TP*AP*CP*AP*TP*A)-3', CcdA | | 著者 | Madl, T, Van Melderen, L, Respondek, M, Oberer, M, Keller, W, Zangger, K. | | 登録日 | 2006-05-22 | | 公開日 | 2006-11-21 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural Basis for Nucleic Acid and Toxin Recognition of the Bacterial Antitoxin CcdA
J.Mol.Biol., 364, 2006
|
|
2JTW
 
 | |
5NMS
 
 | | Hsp21 dodecamer, structural model based on cryo-EM and homology modelling | | 分子名称: | 25.3 kDa heat shock protein, chloroplastic | | 著者 | Rutsdottir, G, Harmark, J, Koeck, P.J.B, Hebert, H, Soderberg, C.A.G, Emanuelsson, C. | | 登録日 | 2017-04-07 | | 公開日 | 2017-05-03 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (10 Å) | | 主引用文献 | Structural model of dodecameric heat-shock protein Hsp21: Flexible N-terminal arms interact with client proteins while C-terminal tails maintain the dodecamer and chaperone activity.
J. Biol. Chem., 292, 2017
|
|
2ADL
 
 | | Solution structure of the bacterial antitoxin CcdA: Implications for DNA and toxin binding | | 分子名称: | CcdA | | 著者 | Madl, T, VanMelderen, L, Oberer, M, Keller, W, Khatai, L, Zangger, K. | | 登録日 | 2005-07-20 | | 公開日 | 2006-08-22 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural basis for nucleic acid and toxin recognition of the bacterial antitoxin CcdA
J.Mol.Biol., 364, 2006
|
|
2ADN
 
 | | Solution structure of the bacterial antitoxin CcdA: Implications for DNA and toxin binding | | 分子名称: | CcdA | | 著者 | Madl, T, VanMelderen, L, Oberer, M, Keller, W, Khatai, L, Zangger, K. | | 登録日 | 2005-07-20 | | 公開日 | 2006-08-22 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural basis for nucleic acid and toxin recognition of the bacterial antitoxin CcdA
J.Mol.Biol., 364, 2006
|
|
3JRZ
 
 | |
3JSC
 
 | | CcdBVfi-FormI-pH7.0 | | 分子名称: | CcdB, SULFATE ION | | 著者 | De Jonge, N, Buts, L, Loris, R. | | 登録日 | 2009-09-10 | | 公開日 | 2009-12-01 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structural and thermodynamic characterization of vibrio fischeri CCDB
J.Biol.Chem., 285, 2010
|
|
2MRN
 
 | |
2MRU
 
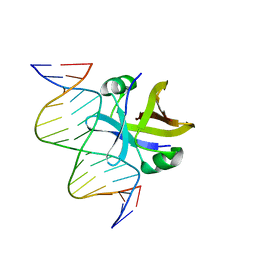 | | Structure of truncated EcMazE-DNA complex | | 分子名称: | Antitoxin MazE, DNA (5'-D(*CP*GP*TP*GP*AP*TP*AP*TP*AP*TP*AP*GP*TP*GP*C)-3'), DNA (5'-D(P*GP*CP*AP*CP*TP*AP*TP*AP*TP*AP*TP*CP*AP*CP*G)-3') | | 著者 | Zorzini, V, Buts, L, Loris, R, van Nuland, N. | | 登録日 | 2014-07-15 | | 公開日 | 2015-02-04 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Escherichia coli antitoxin MazE as transcription factor: insights into MazE-DNA binding.
Nucleic Acids Res., 43, 2015
|
|
2KMT
 
 | |
