6HPD
 
 | | The structure of a beta-glucuronidase from glycoside hydrolase family 2 | | 分子名称: | BROMIDE ION, Beta-galactosidase (GH2), MAGNESIUM ION | | 著者 | Robb, C.S, Gerlach, N, Reisky, L, Bornshoeru, U, Hehemann, J.H. | | 登録日 | 2018-09-20 | | 公開日 | 2019-07-24 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.43 Å) | | 主引用文献 | A marine bacterial enzymatic cascade degrades the algal polysaccharide ulvan.
Nat.Chem.Biol., 15, 2019
|
|
6QG9
 
 | | Crystal structure of Ideonella sakaiensis MHETase | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | 著者 | Palm, G.J, Reisky, L, Boettcher, D, Mueller, H, Michels, E.A.P, Walczak, C, Berndt, L, Weiss, M.S, Bornscheuer, U.T, Weber, G. | | 登録日 | 2019-01-10 | | 公開日 | 2019-04-03 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Structure of the plastic-degrading Ideonella sakaiensis MHETase bound to a substrate.
Nat Commun, 10, 2019
|
|
6QGC
 
 | | PETase from Ideonella sakaiensis without ligand | | 分子名称: | CHLORIDE ION, Poly(ethylene terephthalate) hydrolase, SULFATE ION | | 著者 | Palm, G.J, Reisky, L, Boettcher, D, Mueller, H, Michels, E.A.P, Walczak, C, Berndt, L, Weiss, M.S, Bornscheuer, U.T, Weber, G. | | 登録日 | 2019-01-10 | | 公開日 | 2019-04-03 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure of the plastic-degrading Ideonella sakaiensis MHETase bound to a substrate.
Nat Commun, 10, 2019
|
|
6QGB
 
 | | Crystal structure of Ideonella sakaiensis MHETase bound to benzoic acid | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, BENZOIC ACID, CALCIUM ION, ... | | 著者 | Palm, G.J, Reisky, L, Boettcher, D, Mueller, H, Michels, E.A.P, Walczak, C, Berndt, L, Weiss, M.S, Bornscheuer, U.T, Weber, G. | | 登録日 | 2019-01-10 | | 公開日 | 2019-04-03 | | 最終更新日 | 2019-04-24 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structure of the plastic-degrading Ideonella sakaiensis MHETase bound to a substrate.
Nat Commun, 10, 2019
|
|
6QGA
 
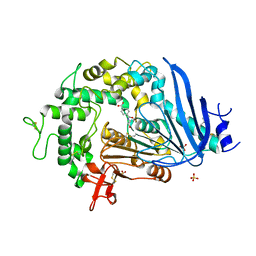 | | Crystal structure of Ideonella sakaiensis MHETase bound to the non-hydrolyzable ligand MHETA | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-(2-hydroxyethylcarbamoyl)benzoic acid, CALCIUM ION, ... | | 著者 | Palm, G.J, Reisky, L, Boettcher, D, Mueller, H, Michels, E.A.P, Walczak, C, Berndt, L, Weiss, M.S, Bornscheuer, U.T, Weber, G. | | 登録日 | 2019-01-10 | | 公開日 | 2019-04-03 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure of the plastic-degrading Ideonella sakaiensis MHETase bound to a substrate.
Nat Commun, 10, 2019
|
|
6HR5
 
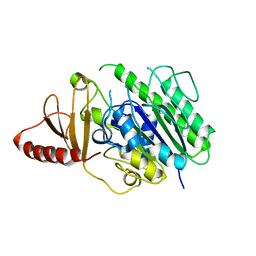 | | Structure of the S1_25 family sulfatase module of the rhamnosidase FA22250 from Formosa agariphila | | 分子名称: | Alpha-L-rhamnosidase/sulfatase (GH78), CALCIUM ION | | 著者 | Roret, T, Prechoux, A, Czjzek, M, Michel, G. | | 登録日 | 2018-09-26 | | 公開日 | 2019-06-26 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (2.912 Å) | | 主引用文献 | A marine bacterial enzymatic cascade degrades the algal polysaccharide ulvan.
Nat.Chem.Biol., 15, 2019
|
|
6HHM
 
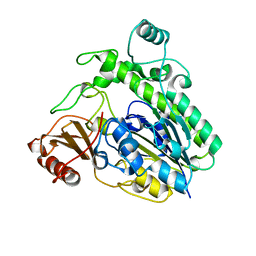 | | Crystal structure of the family S1_7 ulvan-specific sulfatase FA22070 from Formosa agariphila | | 分子名称: | Arylsulfatase, CALCIUM ION | | 著者 | Roret, T, Prechoux, A, Michel, G, Czjzek, M. | | 登録日 | 2018-08-28 | | 公開日 | 2019-06-26 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.23 Å) | | 主引用文献 | A marine bacterial enzymatic cascade degrades the algal polysaccharide ulvan.
Nat.Chem.Biol., 15, 2019
|
|
6HHN
 
 | |
6Y9K
 
 | |
6G5Q
 
 | |
6G5O
 
 | |
