6G81
 
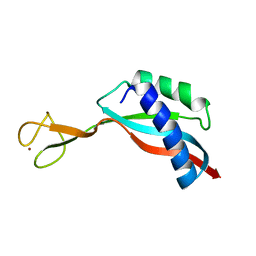 | | Solution structure of the Ni metallochaperone HypA from Helicobacter pylori | | 分子名称: | Hydrogenase maturation factor HypA, ZINC ION | | 著者 | Spronk, C.A.E.M, Zerko, S, Gorka, M, Kozminski, W, Bardiaux, B, Zambelli, B, Musiani, F, Piccioli, M, Hu, H, Maroney, M, Ciurli, S. | | 登録日 | 2018-04-07 | | 公開日 | 2018-10-10 | | 最終更新日 | 2024-06-19 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure and dynamics of Helicobacter pylori nickel-chaperone HypA: an integrated approach using NMR spectroscopy, functional assays and computational tools.
J. Biol. Inorg. Chem., 23, 2018
|
|
1CLF
 
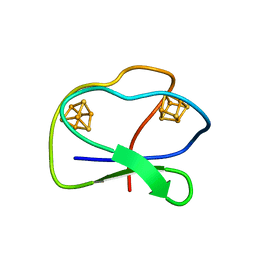 | | CLOSTRIDIUM PASTEURIANUM FERREDOXIN | | 分子名称: | FERREDOXIN, IRON/SULFUR CLUSTER | | 著者 | Bertini, I, Donaire, A, Feinberg, B.A, Luchinat, C, Piccioli, M, Yuan, H. | | 登録日 | 1995-06-21 | | 公開日 | 1996-01-29 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the oxidized 2[4Fe-4S] ferredoxin from Clostridium pasteurianum.
Eur.J.Biochem., 232, 1995
|
|
2K4W
 
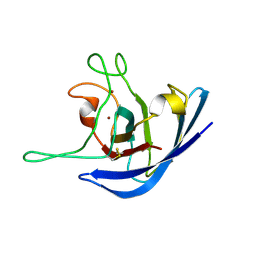 | | The Solution Structure of the Monomeric Copper, Zinc Superoxide Dismutase from Salmonella enterica | | 分子名称: | COPPER (I) ION, Superoxide dismutase [Cu-Zn], ZINC ION | | 著者 | Mori, M, Jimenez, B, Piccioli, M, Battistoni, A, Sette, M, Structural Proteomics in Europe (SPINE) | | 登録日 | 2008-06-20 | | 公開日 | 2008-11-18 | | 最終更新日 | 2021-10-20 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The Solution Structure of the Monomeric Copper, Zinc Superoxide Dismutase from Salmonella enterica: Structural Insights To Understand the Evolution toward the Dimeric Structure.
Biochemistry, 47, 2008
|
|
7A4L
 
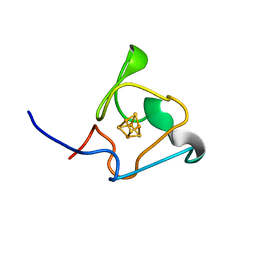 | | PRE-only solution structure of the Iron-Sulfur protein PioC from Rhodopseudomonas palustris TIE-1 | | 分子名称: | IRON/SULFUR CLUSTER, PioC | | 著者 | Trindade, I, Invernici, M, Cantini, F, Louro, R, Piccioli, M. | | 登録日 | 2020-08-19 | | 公開日 | 2020-11-11 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | PRE-driven protein NMR structures: an alternative approach in highly paramagnetic systems.
Febs J., 288, 2021
|
|
2MMZ
 
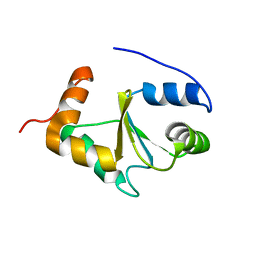 | | Solution structure of the apo form of human glutaredoxin 5 | | 分子名称: | Glutaredoxin-related protein 5, mitochondrial | | 著者 | Banci, L, Brancaccio, D, Ciofi-Baffoni, S, Del Conte, R, Gadepalli, R, Mikolajczyk, M, Neri, S, Piccioli, M, Winkelmann, J. | | 登録日 | 2014-03-25 | | 公開日 | 2014-04-16 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | [2Fe-2S] cluster transfer in iron-sulfur protein biogenesis.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1PIH
 
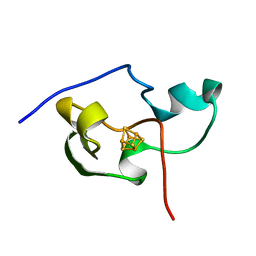 | | THE THREE DIMENSIONAL STRUCTURE OF THE PARAMAGNETIC PROTEIN HIPIP I FROM E.HALOPHILA THROUGH NUCLEAR MAGNETIC RESONANCE | | 分子名称: | HIGH POTENTIAL IRON SULFUR PROTEIN, IRON/SULFUR CLUSTER | | 著者 | Banci, L, Bertini, I, Eltis, L.D, Felli, I, Kastrau, D.H.W, Luchinat, C, Piccioli, M, Pierattelli, R, Smith, M. | | 登録日 | 1994-08-03 | | 公開日 | 1994-12-20 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The three-dimensional structure in solution of the paramagnetic high-potential iron-sulfur protein I from Ectothiorhodospira halophila through nuclear magnetic resonance.
Eur.J.Biochem., 225, 1994
|
|
1PIJ
 
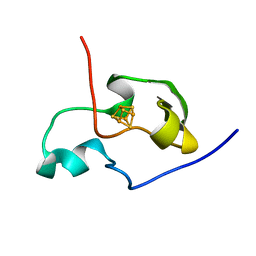 | | THE THREE DIMENSIONAL STRUCTURE OF THE PARAMAGNETIC PROTEIN HIPIP I FROM E.HALOPHILA THROUGH NUCLEAR MAGNETIC RESONANCE | | 分子名称: | HIGH POTENTIAL IRON SULFUR PROTEIN, IRON/SULFUR CLUSTER | | 著者 | Banci, L, Bertini, I, Eltis, L.D, Felli, I.C, Kastrau, D.H.W, Luchinat, C, Piccioli, M, Pierattelli, R, Smith, M. | | 登録日 | 1994-11-11 | | 公開日 | 1995-02-07 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The three-dimensional structure in solution of the paramagnetic high-potential iron-sulfur protein I from Ectothiorhodospira halophila through nuclear magnetic resonance.
Eur.J.Biochem., 225, 1994
|
|
1BA9
 
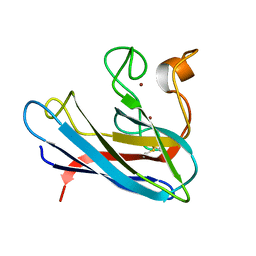 | | THE SOLUTION STRUCTURE OF REDUCED MONOMERIC SUPEROXIDE DISMUTASE, NMR, 36 STRUCTURES | | 分子名称: | COPPER (I) ION, SUPEROXIDE DISMUTASE, ZINC ION | | 著者 | Banci, L, Benedetto, M, Bertini, I, Del Conte, R, Piccioli, M, Viezzoli, M.S. | | 登録日 | 1998-04-24 | | 公開日 | 1998-09-16 | | 最終更新日 | 2021-11-03 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of reduced monomeric Q133M2 copper, zinc superoxide dismutase (SOD). Why is SOD a dimeric enzyme?.
Biochemistry, 37, 1998
|
|
8B9R
 
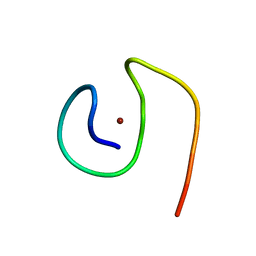 | | Molecular structure of Cu(II)-bound amyloid-beta monomer implicated in inhibition of peptide self-assembly in Alzheimer's disease | | 分子名称: | Amyloid-beta A4 protein, COPPER (II) ION | | 著者 | Abelein, A, Ciofi-Baffoni, S, Morman, C, Kumar, R, Giachetti, A, Piccioli, M, Biverstal, H. | | 登録日 | 2022-10-06 | | 公開日 | 2023-02-01 | | 最終更新日 | 2024-06-19 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Molecular Structure of Cu(II)-Bound Amyloid-beta Monomer Implicated in Inhibition of Peptide Self-Assembly in Alzheimer's Disease.
Jacs Au, 2, 2022
|
|
8B9Q
 
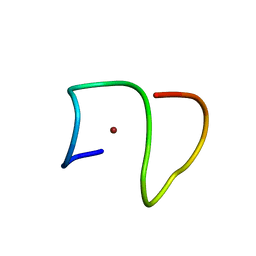 | | Molecular structure of Cu(II)-bound amyloid-beta monomer implicated in inhibition of peptide self-assembly in Alzheimer's disease | | 分子名称: | Amyloid-beta A4 protein, COPPER (II) ION | | 著者 | Abelein, A, Ciofi-Baffoni, S, Kumar, R, Giachetti, A, Piccioli, M, Biverstal, H. | | 登録日 | 2022-10-06 | | 公開日 | 2023-02-08 | | 最終更新日 | 2024-06-05 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Molecular Structure of Cu(II)-Bound Amyloid-beta Monomer Implicated in Inhibition of Peptide Self-Assembly in Alzheimer's Disease.
Jacs Au, 2, 2022
|
|
1N65
 
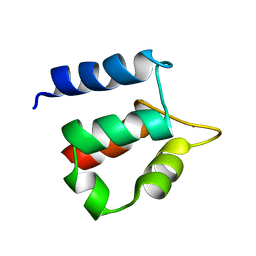 | |
1KSM
 
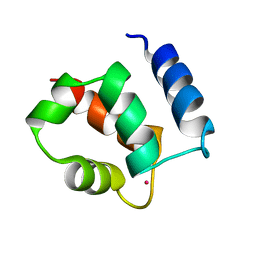 | | AVERAGE NMR SOLUTION STRUCTURE OF CA LN CALBINDIN D9K | | 分子名称: | LANTHANUM (III) ION, VITAMIN D-DEPENDENT CALCIUM-BINDING PROTEIN | | 著者 | Bertini, I, Donaire, A, Luchinat, C, Piccioli, M, Poggi, L, Parigi, G, Jimenez, B. | | 登録日 | 2002-01-14 | | 公開日 | 2002-01-23 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Paramagnetism-based versus classical constraints: an analysis of the solution structure of Ca Ln calbindin D9k.
J.Biomol.NMR, 21, 2001
|
|
1KQV
 
 | | Family of NMR Solution Structures of Ca Ln Calbindin D9K | | 分子名称: | LANTHANUM (III) ION, VITAMIN D-DEPENDENT CALCIUM-BINDING PROTEIN | | 著者 | Bertini, I, Donaire, A, Jimenez, B, Luchinat, C, Parigi, G, Piccioli, M, Poggi, L. | | 登録日 | 2002-01-08 | | 公開日 | 2002-01-16 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Paramagnetism-based versus classical constraints: an analysis of the solution structure of Ca Ln calbindin D9k.
J.Biomol.NMR, 21, 2001
|
|
6XYV
 
 | |
2UYD
 
 | | Crystal structure of the SmHasA mutant H83A | | 分子名称: | ACETATE ION, HEMOPHORE HASA, PROTOPORPHYRIN IX CONTAINING FE, ... | | 著者 | Czjzek, M, Caillet-Saguy, C, Fournelle, A, Guigliarelli, B, Izadi-Pruneyre, N, Lecroisey, A. | | 登録日 | 2007-04-04 | | 公開日 | 2007-12-25 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Deciphering the Structural Role of Histidine 83 for Heme Binding in Hemophore Hasa.
J.Biol.Chem., 283, 2008
|
|
7A58
 
 | |
4H2D
 
 | | Crystal structure of NDOR1 | | 分子名称: | FLAVIN MONONUCLEOTIDE, NADPH-dependent diflavin oxidoreductase 1 | | 著者 | Banci, L, Bertini, I, Calderone, V, Ciofi-Baffoni, S, Mikolajczyk, M, Jaiswal, D, Winkelmann, J. | | 登録日 | 2012-09-12 | | 公開日 | 2013-04-17 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Molecular view of an electron transfer process essential for iron-sulfur protein biogenesis.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
7QP5
 
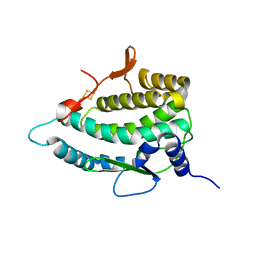 | | Crystal Structure of E. coli FhuF | | 分子名称: | FE2/S2 (INORGANIC) CLUSTER, Ferric iron reductase protein FhuF | | 著者 | Trindade, I.B, Rollo, F, Matias, P.M, Moe, E, Louro, R.O. | | 登録日 | 2022-01-03 | | 公開日 | 2023-07-12 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | The structure of a novel ferredoxin: FhuF, a ferric-siderophore reductase from E. coli K-12 with a novel 2Fe-2S cluster coordination
Biorxiv, 2023
|
|
1NEH
 
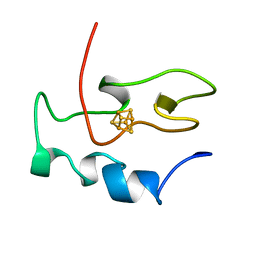 | | HIGH POTENTIAL IRON-SULFUR PROTEIN | | 分子名称: | HIGH POTENTIAL IRON SULFUR PROTEIN, IRON/SULFUR CLUSTER | | 著者 | Bertini, I, Dikiy, A, Kastrau, D.H.W, Luchinat, C, Sompornpisut, P. | | 登録日 | 1995-12-14 | | 公開日 | 1996-03-08 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Three-dimensional solution structure of the oxidized high potential iron-sulfur protein from Chromatium vinosum through NMR. Comparative analysis with the solution structure of the reduced species.
Biochemistry, 34, 1995
|
|
1HRQ
 
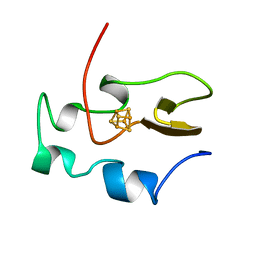 | | THE THREE-DIMENSIONAL SOLUTION STRUCTURE OF THE REDUCED HIGH-POTENTIAL IRON-SULFUR PROTEIN FROM CHROMATIUM VINOSUM THROUGH NMR | | 分子名称: | HIGH POTENTIAL IRON SULFUR PROTEIN, IRON/SULFUR CLUSTER | | 著者 | Banci, L, Bertini, I, Dikiy, A, Kastrau, D.H.W, Luchinat, C, Sompornpisut, P. | | 登録日 | 1995-01-17 | | 公開日 | 1995-06-03 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The three-dimensional solution structure of the reduced high-potential iron-sulfur protein from Chromatium vinosum through NMR.
Biochemistry, 34, 1995
|
|
1HRR
 
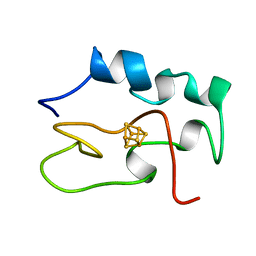 | | THE THREE DIMENSIONAL STRUCTURE OF THE REDUCED HIGH POTENTIAL IRON-SULFUR PROTEIN FROM CHROMATIUM VINOSUM THROUGH NMR | | 分子名称: | IRON/SULFUR CLUSTER, REDUCED HIGH POTENTIAL IRON SULFUR PROTEIN | | 著者 | Banci, L, Bertini, I, Dikiy, A, Kastrau, D.H.W, Luchinat, C, Sompornpisut, P. | | 登録日 | 1995-01-17 | | 公開日 | 1995-07-31 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The three-dimensional solution structure of the reduced high-potential iron-sulfur protein from Chromatium vinosum through NMR.
Biochemistry, 34, 1995
|
|
