5FRV
 
 | |
5FS0
 
 | |
5FRX
 
 | |
5FRY
 
 | |
5FRZ
 
 | |
5FRU
 
 | |
5FRW
 
 | |
5GN3
 
 | |
5GRK
 
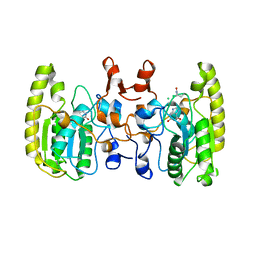 | | Crystal structure of Uracil DNA glycosylase -Xanthine complex from Bradyrhizobium diazoefficiens | | 分子名称: | Blr0248 protein, XANTHINE | | 著者 | Patil, V.V, Ullas, V.C, Ahn, W, Varshney, U, Woo, E. | | 登録日 | 2016-08-11 | | 公開日 | 2017-05-03 | | 最終更新日 | 2017-06-14 | | 実験手法 | X-RAY DIFFRACTION (2.804 Å) | | 主引用文献 | Uracil DNA glycosylase (UDG) activities in Bradyrhizobium diazoefficiens: characterization of a new class of UDG with broad substrate specificity
Nucleic Acids Res., 45, 2017
|
|
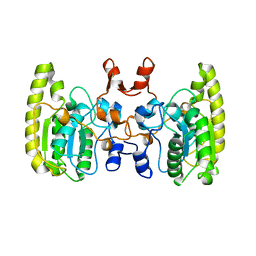
5GN2
 
 | |
5GNW
 
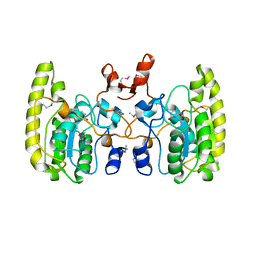 | | Crystal structure of Uracil DNA glycosylase-Uracil complex from Bradyrhizobium diazoefficiens. | | 分子名称: | Blr0248 protein, URACIL | | 著者 | Patil, V.V, Chembazhi, U.V, Varshney, U, Woo, E. | | 登録日 | 2016-07-25 | | 公開日 | 2017-06-14 | | 実験手法 | X-RAY DIFFRACTION (2.869 Å) | | 主引用文献 | Uracil DNA glycosylase (UDG) activities in Bradyrhizobium diazoefficiens: characterization of a new class of UDG with broad substrate specificity
Nucleic Acids Res., 45, 2017
|
|
6IY8
 
 | | DmpR-phenol complex of Pseudomonas putida | | 分子名称: | PHENOL, Positive regulator CapR, ZINC ION | | 著者 | Park, K.H, Woo, E.J. | | 登録日 | 2018-12-13 | | 公開日 | 2020-06-10 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (3.42 Å) | | 主引用文献 | Tetrameric architecture of an active phenol-bound form of the AAA+transcriptional regulator DmpR.
Nat Commun, 11, 2020
|
|
