1P8L
 
 | | New Crystal Structure of Chlorella Virus DNA Ligase-Adenylate | | 分子名称: | ADENOSINE MONOPHOSPHATE, PBCV-1 DNA ligase | | 著者 | Odell, M, Malinina, L, Teplova, M, Shuman, S. | | 登録日 | 2003-05-07 | | 公開日 | 2003-08-26 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.95 Å) | | 主引用文献 | Analysis of the DNA Joining Repertoire of Chlorella Virus DNA ligase and a New Crystal Structure of the Ligase-Adenylate Intermediate
Nucleic Acids Res., 31, 2003
|
|
1FVI
 
 | | CRYSTAL STRUCTURE OF CHLORELLA VIRUS DNA LIGASE-ADENYLATE | | 分子名称: | ADENOSINE MONOPHOSPHATE, CHLORELLA VIRUS DNA LIGASE-ADENYLATE, SULFATE ION | | 著者 | Odell, M, Sriskanda, V, Shuman, S, Nikolov, D.B. | | 登録日 | 2000-09-20 | | 公開日 | 2000-11-22 | | 最終更新日 | 2021-11-03 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of eukaryotic DNA ligase-adenylate illuminates the mechanism of nick sensing and strand joining.
Mol.Cell, 6, 2000
|
|
4BTE
 
 | | DJ-1 Cu(I) complex | | 分子名称: | COPPER (I) ION, PROTEIN DJ-1 | | 著者 | Puno, M.R.A, Odell, M, Moody, P.C.E. | | 登録日 | 2013-06-14 | | 公開日 | 2013-11-06 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.38 Å) | | 主引用文献 | Structure of Cu(I)-Bound Dj-1 Reveals a Biscysteinate Metal Binding Site at the Homodimer Interface: Insights Into Mutational Inactivation of Dj-1 in Parkinsonism.
J.Am.Chem.Soc., 135, 2013
|
|
1DUS
 
 | | MJ0882-A hypothetical protein from M. jannaschii | | 分子名称: | MJ0882 | | 著者 | Hung, L, Huang, L, Kim, R, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | 登録日 | 2000-01-18 | | 公開日 | 2000-07-19 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structure-based experimental confirmation of biochemical function to a methyltransferase, MJ0882, from hyperthermophile Methanococcus jannaschii
J.STRUCT.FUNCT.GENOM., 2, 2002
|
|
2Q2T
 
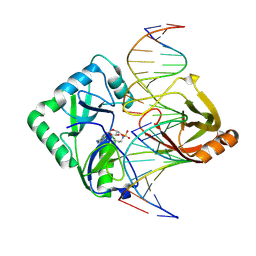 | | Structure of Chlorella virus DNA ligase-adenylate bound to a 5' phosphorylated nick | | 分子名称: | 5'-D(*AP*TP*TP*GP*CP*GP*AP*CP*(OMC)P*C)-3', 5'-D(*TP*TP*CP*CP*GP*AP*TP*AP*GP*TP*GP*GP*GP*GP*TP*CP*GP*CP*AP*AP*T)-3', 5'-D(P*CP*AP*CP*TP*AP*TP*CP*GP*GP*AP*A)-3', ... | | 著者 | Lima, C.D, Nandakumar, J, Nair, P.A, Smith, P, Shuman, S. | | 登録日 | 2007-05-29 | | 公開日 | 2007-07-10 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural basis for nick recognition by a minimal pluripotent DNA ligase.
Nat.Struct.Mol.Biol., 14, 2007
|
|
2Q2U
 
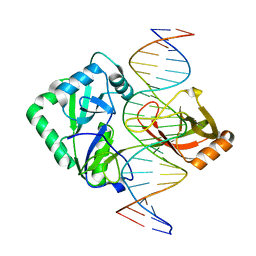 | | Structure of Chlorella virus DNA ligase-product DNA complex | | 分子名称: | 5'-D(*AP*TP*TP*GP*CP*GP*AP*CP*(OMC)P*CP*CP*AP*CP*TP*AP*TP*CP*GP*GP*AP*A)-3', 5'-D(*TP*TP*CP*CP*GP*AP*TP*AP*GP*TP*GP*GP*GP*GP*TP*CP*GP*CP*AP*AP*T)-3', Chlorella virus DNA ligase | | 著者 | Lima, C.D, Nandakumar, J, Nair, P.A, Smith, P, Shuman, S. | | 登録日 | 2007-05-29 | | 公開日 | 2007-07-10 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structural basis for nick recognition by a minimal pluripotent DNA ligase.
Nat.Struct.Mol.Biol., 14, 2007
|
|
