6XC0
 
 | | Crystal structure of bacteriophage T4 spackle and lysozyme in monoclinic form | | 分子名称: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | 著者 | Shi, K, Oakland, J.T, Kurniawan, F, Moeller, N.H, Aihara, H. | | 登録日 | 2020-06-07 | | 公開日 | 2020-12-02 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | Structural basis of superinfection exclusion by bacteriophage T4 Spackle.
Commun Biol, 3, 2020
|
|
6XC1
 
 | | Crystal structure of bacteriophage T4 spackle and lysozyme in orthorhombic form | | 分子名称: | 1,2-ETHANEDIOL, ISOPROPYL ALCOHOL, Lysozyme, ... | | 著者 | Shi, K, Oakland, J.T, Kurniawan, F, Moeller, N.H, Aihara, H. | | 登録日 | 2020-06-07 | | 公開日 | 2020-12-02 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | Structural basis of superinfection exclusion by bacteriophage T4 Spackle.
Commun Biol, 3, 2020
|
|
6X6O
 
 | | Crystal structure of T4 protein Spackle as determined by native SAD phasing | | 分子名称: | CHLORIDE ION, Protein spackle | | 著者 | Shi, K, Kurniawan, F, Banerjee, S, Moeller, N.H, Aihara, H. | | 登録日 | 2020-05-28 | | 公開日 | 2020-09-16 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (1.52 Å) | | 主引用文献 | Crystal structure of bacteriophage T4 Spackle as determined by native SAD phasing.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
7MC6
 
 | | Crystal structure of the SARS-CoV-2 ExoN-nsp10 complex containing Mg2+ ion | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, MAGNESIUM ION, ... | | 著者 | Moeller, N.M, Shi, K, Banerjee, S, Yin, L, Aihara, H. | | 登録日 | 2021-04-01 | | 公開日 | 2021-05-05 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure and dynamics of SARS-CoV-2 proofreading exoribonuclease ExoN.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7MC5
 
 | | Crystal structure of the SARS-CoV-2 ExoN-nsp10 complex | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, L(+)-TARTARIC ACID, ... | | 著者 | Moeller, N.M, Shi, K, Banerjee, S, Yin, L, Aihara, H. | | 登録日 | 2021-04-01 | | 公開日 | 2021-05-05 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.64 Å) | | 主引用文献 | Structure and dynamics of SARS-CoV-2 proofreading exoribonuclease ExoN.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7SPP
 
 | |
7SPO
 
 | |
7K32
 
 | | Crystal structure of Endonuclease Q complex with 27-mer duplex substrate with an abasic lesion at the active site | | 分子名称: | DNA (27-MER), Endonuclease Q, MAGNESIUM ION, ... | | 著者 | Shi, K, Moeller, N.M, Banerjee, S, Yin, L, Orellana, K, Aihara, H. | | 登録日 | 2020-09-10 | | 公開日 | 2021-03-17 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.11 Å) | | 主引用文献 | Structural basis for recognition of distinct deaminated DNA lesions by endonuclease Q.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7K33
 
 | | Crystal structure of Endonuclease Q complex with 27-mer duplex substrate with an abasic lesion at the active site | | 分子名称: | DNA (27-MER), Endonuclease Q, MAGNESIUM ION, ... | | 著者 | Shi, K, Moeller, N.M, Banerjee, S, Yin, L, Orellana, K, Aihara, H. | | 登録日 | 2020-09-10 | | 公開日 | 2021-03-17 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.11 Å) | | 主引用文献 | Structural basis for recognition of distinct deaminated DNA lesions by endonuclease Q.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7K30
 
 | | Crystal structure of Endonuclease Q complex with 27-mer duplex substrate with dU at the active site | | 分子名称: | 1,2-ETHANEDIOL, DNA (27-MER), Endonuclease Q, ... | | 著者 | Shi, K, Moeller, N.M, Banerjee, S, Yin, L, Orellana, K, Aihara, H. | | 登録日 | 2020-09-10 | | 公開日 | 2021-03-17 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.34 Å) | | 主引用文献 | Structural basis for recognition of distinct deaminated DNA lesions by endonuclease Q.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7K31
 
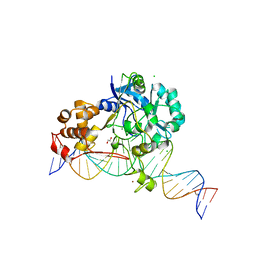 | | Crystal structure of Endonuclease Q complex with 27-mer duplex substrate with dI at the active site | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA (27-MER), ... | | 著者 | Shi, K, Moeller, N.M, Banerjee, S, Yin, L, Orellana, K, Aihara, H. | | 登録日 | 2020-09-10 | | 公開日 | 2021-03-17 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.88 Å) | | 主引用文献 | Structural basis for recognition of distinct deaminated DNA lesions by endonuclease Q.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6VOY
 
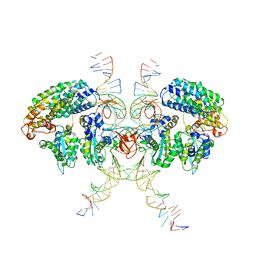 | | Cryo-EM structure of HTLV-1 instasome | | 分子名称: | DNA (25-MER), DNA (5'-D(P*AP*CP*AP*CP*AP*CP*TP*TP*GP*AP*CP*TP*AP*GP*GP*GP*TP*G)-3'), DNA-binding protein 7d, ... | | 著者 | Bhatt, V, Shi, K, Sundborger, A, Aihara, H. | | 登録日 | 2020-02-01 | | 公開日 | 2020-07-01 | | 最終更新日 | 2024-10-09 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Structural basis of host protein hijacking in human T-cell leukemia virus integration.
Nat Commun, 11, 2020
|
|
8V9Z
 
 | |
8V9Y
 
 | |
8VA0
 
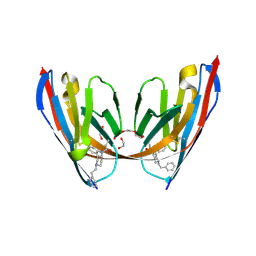 | | X-ray crystal structure of JGFN4 N76D complexed with fentanyl in dimer form | | 分子名称: | 1,2-ETHANEDIOL, JGFN4, N-phenyl-N-[1-(2-phenylethyl)piperidin-4-yl]propanamide, ... | | 著者 | Shi, K, Moller, N, Aihara, H. | | 登録日 | 2023-12-10 | | 公開日 | 2024-08-07 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Identification and biophysical characterization of a novel domain-swapped camelid antibody specific for fentanyl.
J.Biol.Chem., 300, 2024
|
|
8V9X
 
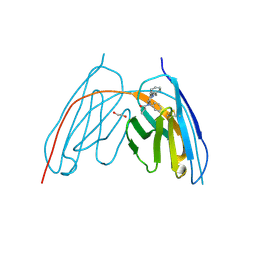 | | X-ray crystal structure of JGFN4 complex with fentanyl | | 分子名称: | 1,2-ETHANEDIOL, JGFN4, N-phenyl-N-[1-(2-phenylethyl)piperidin-4-yl]propanamide | | 著者 | Moller, N, Shi, K, Aihara, H. | | 登録日 | 2023-12-10 | | 公開日 | 2024-08-07 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Identification and biophysical characterization of a novel domain-swapped camelid antibody specific for fentanyl.
J.Biol.Chem., 300, 2024
|
|
8V9W
 
 | | X-ray crystal structure of JGFN4 complexed with fentanyl | | 分子名称: | 1,2-ETHANEDIOL, JGFN4, N-phenyl-N-[1-(2-phenylethyl)piperidin-4-yl]propanamide | | 著者 | Moller, N, Shi, K, Aihara, H. | | 登録日 | 2023-12-10 | | 公開日 | 2024-08-07 | | 実験手法 | X-RAY DIFFRACTION (1.68 Å) | | 主引用文献 | Identification and biophysical characterization of a novel domain-swapped camelid antibody specific for fentanyl.
J.Biol.Chem., 300, 2024
|
|
7KM6
 
 | | APOBEC3B antibody 5G7 Fv-clasp | | 分子名称: | 1,2-ETHANEDIOL, 5G7 human monoclonal FAB heavy chain, 5G7 human monoclonal FAB light chain, ... | | 著者 | Tang, H, Shi, K, Aihara, H. | | 登録日 | 2020-11-02 | | 公開日 | 2021-05-12 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (1.67 Å) | | 主引用文献 | Structural Characterization of a Minimal Antibody against Human APOBEC3B.
Viruses, 13, 2021
|
|
