7DFS
 
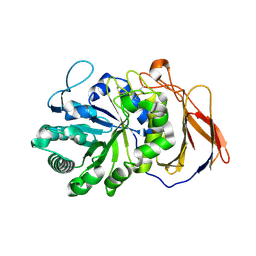 | | Crystal structure of a novel 4-O-alpha-L-rhamnosyl-beta-D-glucuronidase from Fusarium oxysporum 12S - Rha-GlcA complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-O-alpha-L-rhamnosyl-beta-D-glucuronidase, alpha-D-mannopyranose, ... | | 著者 | Kondo, T, Arakawa, T, Fushinobu, S, Sakamoto, T. | | 登録日 | 2020-11-09 | | 公開日 | 2021-03-17 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.49 Å) | | 主引用文献 | Biochemical and structural characterization of a novel 4-O-alpha-l-rhamnosyl-beta-d-glucuronidase from Fusarium oxysporum.
Febs J., 288, 2021
|
|
7DFQ
 
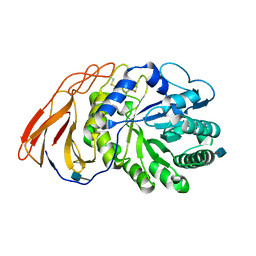 | | Crystal Structure of a novel 4-O-alpha-L-rhamnosyl-beta-D-glucuronidase from Fusarium oxysporum 12S, ligand-free form | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-O-alpha-L-rhamnosyl-beta-D-glucuronidase | | 著者 | Kondo, T, Arakawa, T, Fushinobu, S, Sakamoto, T. | | 登録日 | 2020-11-09 | | 公開日 | 2021-03-17 | | 最終更新日 | 2021-08-25 | | 実験手法 | X-RAY DIFFRACTION (1.51 Å) | | 主引用文献 | Biochemical and structural characterization of a novel 4-O-alpha-l-rhamnosyl-beta-d-glucuronidase from Fusarium oxysporum.
Febs J., 288, 2021
|
|
7ESM
 
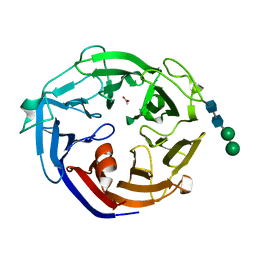 | | Crystal structure of a L-rhamnose-alpha-1,4-D-glucuronate lyase from Fusarium oxysporum 12S, L-Rha complex | | 分子名称: | ACETATE ION, L-rhamnose-alpha-1,4-D-glucuronate lyase, SODIUM ION, ... | | 著者 | Kondo, T, Arakawa, T, Fushinobu, S, Sakamoto, T. | | 登録日 | 2021-05-11 | | 公開日 | 2021-08-04 | | 最終更新日 | 2021-09-01 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Structural and functional analysis of gum arabic l-rhamnose-alpha-1,4-d-glucuronate lyase establishes a novel polysaccharide lyase family.
J.Biol.Chem., 297, 2021
|
|
7ESN
 
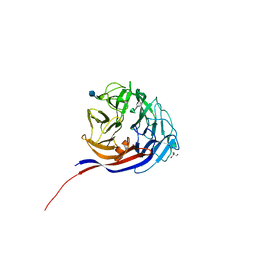 | | Crystal structure of a L-rhamnose-alpha-1,4-D-glucuronate lyase from Fusarium oxysporum 12S, H105F Rha-GlcA complex | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, L-Rhamnose-alpha-1,4-D-glucuronate lyase, ... | | 著者 | Kondo, T, Arakawa, T, Fushinobu, S, Sakamoto, T. | | 登録日 | 2021-05-11 | | 公開日 | 2021-08-04 | | 最終更新日 | 2021-09-01 | | 実験手法 | X-RAY DIFFRACTION (2.42 Å) | | 主引用文献 | Structural and functional analysis of gum arabic l-rhamnose-alpha-1,4-d-glucuronate lyase establishes a novel polysaccharide lyase family.
J.Biol.Chem., 297, 2021
|
|
7ESK
 
 | | Crystal structure of a L-rhamnose-alpha-1,4-D-glucuronate lyase from Fusarium oxysporum 12S, Ligand free form | | 分子名称: | CALCIUM ION, L-Rhamnose-alpha-1,4-D-glucuronate lyase, SODIUM ION, ... | | 著者 | Kondo, T, Arakawa, T, Fushinobu, S, Sakamoto, T. | | 登録日 | 2021-05-11 | | 公開日 | 2021-08-04 | | 最終更新日 | 2021-09-01 | | 実験手法 | X-RAY DIFFRACTION (1.05 Å) | | 主引用文献 | Structural and functional analysis of gum arabic l-rhamnose-alpha-1,4-d-glucuronate lyase establishes a novel polysaccharide lyase family.
J.Biol.Chem., 297, 2021
|
|
7ESL
 
 | | Crystal structure of a L-rhamnose-alpha-1,4-D-glucuronate lyase from Fusarium oxysporum 12S, N247A N-glycan free form | | 分子名称: | L-rhamnose-alpha-1,4-D-glucuronate lyase, SODIUM ION | | 著者 | Kondo, T, Arakawa, T, Fushinobu, S, Sakamoto, T. | | 登録日 | 2021-05-11 | | 公開日 | 2021-08-04 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Structural and functional analysis of gum arabic l-rhamnose-alpha-1,4-d-glucuronate lyase establishes a novel polysaccharide lyase family.
J.Biol.Chem., 297, 2021
|
|
8I4D
 
 | | X-ray structure of a L-rhamnose-alpha-1,4-D-glucuronate lyase from Fusarium oxysporum 12S, L-Rha complex at 100K | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, CALCIUM ION, ... | | 著者 | Yano, N, Kondo, T, Kusaka, K, Yamada, T, Arakawa, T, Sakamoto, T, Fushinobu, S. | | 登録日 | 2023-01-19 | | 公開日 | 2024-01-24 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.06 Å) | | 主引用文献 | Charge neutralization and beta-elimination cleavage mechanism of family 42 L-rhamnose-alpha-1,4-D-glucuronate lyase revealed using neutron crystallography.
J.Biol.Chem., 300, 2024
|
|
7YQS
 
 | | Neutron structure of a L-rhamnose-alpha-1,4-D-glucuronate lyase from Fusarium oxysporum 12S, L-Rha complex | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, CALCIUM ION, ... | | 著者 | Yano, N, Kondo, T, Kusaka, K, Yamada, T, Arakawa, T, Sakamoto, T, Fushinobu, S. | | 登録日 | 2022-08-08 | | 公開日 | 2023-08-09 | | 最終更新日 | 2024-03-27 | | 実験手法 | NEUTRON DIFFRACTION (1.25 Å), X-RAY DIFFRACTION | | 主引用文献 | Charge neutralization and beta-elimination cleavage mechanism of family 42 L-rhamnose-alpha-1,4-D-glucuronate lyase revealed using neutron crystallography.
J.Biol.Chem., 300, 2024
|
|
6IK8
 
 | | Crystal structure of tomato beta-galactosidase (TBG) 4 in complex with beta-1,6-galactobiose | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-galactosidase, ... | | 著者 | Matsuyama, K, Nakae, S, Igarashi, K, Tada, T, Ishimaru, M. | | 登録日 | 2018-10-15 | | 公開日 | 2018-11-28 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Substrate-recognition mechanism of tomato beta-galactosidase 4 using X-ray crystallography and docking simulation.
Planta, 252, 2020
|
|
6IK5
 
 | | Crystal structure of tomato beta-galactosidase (TBG) 4 in complex with galactose | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-galactosidase, ... | | 著者 | Matsuyama, K, Nakae, S, Igarashi, K, Tada, T, Ishimaru, M. | | 登録日 | 2018-10-15 | | 公開日 | 2018-11-28 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.82 Å) | | 主引用文献 | Substrate-recognition mechanism of tomato beta-galactosidase 4 using X-ray crystallography and docking simulation.
Planta, 252, 2020
|
|
6IK7
 
 | | Crystal structure of tomato beta-galactosidase (TBG) 4 in complex with beta-1,3-galactobiose | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-galactosidase, ... | | 著者 | Matsuyama, K, Nakae, S, Igarashi, K, Tada, T, Ishimaru, M. | | 登録日 | 2018-10-15 | | 公開日 | 2018-11-28 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Substrate-recognition mechanism of tomato beta-galactosidase 4 using X-ray crystallography and docking simulation.
Planta, 252, 2020
|
|
6IK6
 
 | | Crystal structure of Tomato beta-galactosidase (TBG) 4 with beta-1,4-galactobiose | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-galactosidase, ... | | 著者 | Matsuyama, K, Nakae, S, Igarashi, K, Tada, T, Ishimaru, M. | | 登録日 | 2018-10-15 | | 公開日 | 2018-11-28 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.791 Å) | | 主引用文献 | Substrate-recognition mechanism of tomato beta-galactosidase 4 using X-ray crystallography and docking simulation.
Planta, 252, 2020
|
|
4TLE
 
 | | Crystal structure of N-terminal C1 domain of KaiC | | 分子名称: | CHLORIDE ION, Circadian clock protein kinase KaiC, MAGNESIUM ION, ... | | 著者 | Abe, J, Hiyama, T.B, Mukaiyama, A, Son, S, Akiyama, S. | | 登録日 | 2014-05-29 | | 公開日 | 2015-07-01 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.936 Å) | | 主引用文献 | Atomic-scale origins of slowness in the cyanobacterial circadian clock
Science, 349, 2015
|
|
4TL7
 
 | | Crystal structure of N-terminal C1 domain of KaiC | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, Circadian clock protein kinase KaiC, ... | | 著者 | Abe, J, Hiyama, T.B, Mukaiyama, A, Son, S, Akiyama, S. | | 登録日 | 2014-05-29 | | 公開日 | 2015-07-01 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.936 Å) | | 主引用文献 | Circadian rhythms. Atomic-scale origins of slowness in the cyanobacterial circadian clock.
Science, 349, 2015
|
|
4TLC
 
 | | Crystal structure of N-terminal C1 domain of KaiC | | 分子名称: | CHLORIDE ION, Circadian clock protein kinase KaiC, MAGNESIUM ION, ... | | 著者 | Abe, J, Hiyama, T.B, Mukaiyama, A, Son, S, Akiyama, S. | | 登録日 | 2014-05-29 | | 公開日 | 2015-07-01 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.09 Å) | | 主引用文献 | Atomic-scale origins of slowness in the cyanobacterial circadian clock
Science, 349, 2015
|
|
4TLB
 
 | | Crystal structure of N-terminal C1 domain of KaiC | | 分子名称: | CHLORIDE ION, Circadian clock protein kinase KaiC, MAGNESIUM ION, ... | | 著者 | Abe, J, Hiyama, T.B, Mukaiyama, A, Son, S, Akiyama, S. | | 登録日 | 2014-05-29 | | 公開日 | 2015-07-01 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.983 Å) | | 主引用文献 | Circadian rhythms. Atomic-scale origins of slowness in the cyanobacterial circadian clock.
Science, 349, 2015
|
|
4TLA
 
 | | Crystal structure of N-terminal C1 domain of KaiC | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, Circadian clock protein kinase KaiC, ... | | 著者 | Abe, J, Hiyama, T.B, Mukaiyama, A, Son, S, Akiyama, S. | | 登録日 | 2014-05-29 | | 公開日 | 2015-07-01 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Circadian rhythms. Atomic-scale origins of slowness in the cyanobacterial circadian clock.
Science, 349, 2015
|
|
4TL9
 
 | | Crystal structure of N-terminal C1 domain of KaiC | | 分子名称: | CHLORIDE ION, Circadian clock protein kinase KaiC, MAGNESIUM ION, ... | | 著者 | Abe, J, Hiyama, T.B, Mukaiyama, A, Son, S, Akiyama, S. | | 登録日 | 2014-05-29 | | 公開日 | 2015-07-01 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.822 Å) | | 主引用文献 | Circadian rhythms. Atomic-scale origins of slowness in the cyanobacterial circadian clock.
Science, 349, 2015
|
|
4TL8
 
 | | Crystal structure of N-terminal C1 domain of KaiC | | 分子名称: | CHLORIDE ION, Circadian clock protein kinase KaiC, MAGNESIUM ION, ... | | 著者 | Abe, J, Hiyama, T.B, Mukaiyama, A, Son, S, Akiyama, S. | | 登録日 | 2014-05-29 | | 公開日 | 2015-07-01 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.859 Å) | | 主引用文献 | Atomic-scale origins of slowness in the cyanobacterial circadian clock
Science, 349, 2015
|
|
4TL6
 
 | | Crystal structure of N-terminal domain of KaiC | | 分子名称: | CHLORIDE ION, Circadian clock protein kinase KaiC, MAGNESIUM ION, ... | | 著者 | Abe, J, Hiyama, T.B, Mukaiyama, A, Son, S, Akiyama, S. | | 登録日 | 2014-05-29 | | 公開日 | 2015-07-01 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.763 Å) | | 主引用文献 | Circadian rhythms. Atomic-scale origins of slowness in the cyanobacterial circadian clock.
Science, 349, 2015
|
|
4TLD
 
 | | Crystal structure of N-terminal C1 domain of KaiC | | 分子名称: | CHLORIDE ION, Circadian clock protein kinase KaiC, MAGNESIUM ION, ... | | 著者 | Abe, J, Hiyama, T.B, Mukaiyama, A, Son, S, Akiyama, S. | | 登録日 | 2014-05-29 | | 公開日 | 2015-07-01 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.949 Å) | | 主引用文献 | Atomic-scale origins of slowness in the cyanobacterial circadian clock
Science, 349, 2015
|
|
7DYE
 
 | |
7DY1
 
 | |
5YZ8
 
 | | Crystal Structure of N-terminal C1 domain of KaiC | | 分子名称: | CHLORIDE ION, Circadian Clock Protein Kinase KaiC, MAGNESIUM ION, ... | | 著者 | Mukaiyama, A, Furuike, Y, Abe, J, Akiyama, S. | | 登録日 | 2017-12-13 | | 公開日 | 2019-01-30 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.81 Å) | | 主引用文献 | Conformational rearrangements of the C1 ring in KaiC measure the timing of assembly with KaiB.
Sci Rep, 8, 2018
|
|
7V3X
 
 | |
