1MXJ
 
 | | NMR solution structure of benz[a]anthracene-dG in ras codon 12,2; GGCAGXTGGTG | | 分子名称: | 1S,2R,3S,4R-TETRAHYDRO-BENZO[A]ANTHRACENE-2,3,4-TRIOL, 5'-D(*CP*AP*CP*CP*AP*CP*CP*TP*GP*CP*C)-3', 5'-D(*GP*GP*CP*AP*GP*GP*TP*GP*GP*TP*G)-3' | | 著者 | Kim, H.-Y.H, Wilkinson, A.S, Harris, C.M, Harris, T.M, Stone, M.P. | | 登録日 | 2002-10-02 | | 公開日 | 2003-03-11 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Minor Groove Orientation for the (1S,2R,3S,4R)-N2-[1-(1,2,3,4-tetrahydro-2,3,4-trihydroxy-benz[a]anthracenyl)]-2'-deoxyguanosyl Adduct in the N-ras Codon 12 sequence
Biochemistry, 42, 2003
|
|
5HKN
 
 | | Crystal structure de novo designed fullerene organizing protein complex with fullerene | | 分子名称: | (C_{60}-I_{h})[5,6]fullerene, fullerene organizing protein | | 著者 | Paul, J, Acharya, R, Kim, K.-H, Kim, Y.H, Kim, N.H, Grigoryan, G, DeGardo, W.F. | | 登録日 | 2016-01-14 | | 公開日 | 2016-05-04 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.761 Å) | | 主引用文献 | Protein-directed self-assembly of a fullerene crystal.
Nat Commun, 7, 2016
|
|
5HKR
 
 | | Crystal structure of de novo designed fullerene organising protein complex with fullerene | | 分子名称: | (C_{60}-I_{h})[5,6]fullerene, fullerene organizing protein | | 著者 | Acharya, R, Kim, Y.H, Grigoryan, G, DeGardo, W.F. | | 登録日 | 2016-01-14 | | 公開日 | 2016-05-04 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Protein-directed self-assembly of a fullerene crystal.
Nat Commun, 7, 2016
|
|
5ET3
 
 | | Crystal Structure of De novo Designed Fullerene organizing peptide | | 分子名称: | (C_{60}-I_{h})[5,6]fullerene, Fullerene Organizing Protein (C60Sol-COP-3) | | 著者 | Kim, K.-H, Kim, Y.H, Acharya, R, Kim, N.H, Paul, J, Grigoryan, G, DeGrado, W.F. | | 登録日 | 2015-11-17 | | 公開日 | 2016-05-04 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.671 Å) | | 主引用文献 | Protein-directed self-assembly of a fullerene crystal.
Nat Commun, 7, 2016
|
|
3S0R
 
 | |
5YU4
 
 | | Structural basis for recognition of L-lysine, L-ornithine, and L-2,4-diamino butyric acid by lysine cyclodeaminase | | 分子名称: | 2,4-DIAMINOBUTYRIC ACID, Lysine cyclodeaminase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | 著者 | Min, K.J, Yoon, H.J, Matsuura, A, Kim, Y.H, Lee, H.H. | | 登録日 | 2017-11-20 | | 公開日 | 2018-05-02 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.144 Å) | | 主引用文献 | Structural Basis for Recognition of L-lysine, L-ornithine, and L-2,4-diamino Butyric Acid by Lysine Cyclodeaminase.
Mol. Cells, 41, 2018
|
|
5YU0
 
 | | Structural basis for recognition of L-lysine, L-ornithine, and L-2,4-diamino butyric acid by lysine cyclodeaminase | | 分子名称: | Lysine cyclodeaminase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION | | 著者 | Min, K.J, Yoon, H.J, Matsuura, A, Kim, Y.H, Lee, H.H. | | 登録日 | 2017-11-20 | | 公開日 | 2018-05-02 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | Structural Basis for Recognition of L-lysine, L-ornithine, and L-2,4-diamino Butyric Acid by Lysine Cyclodeaminase.
Mol. Cells, 41, 2018
|
|
5YU1
 
 | | Structural basis for recognition of L-lysine, L-ornithine, and L-2,4-diamino butyric acid by lysine cyclodeaminase | | 分子名称: | (2S)-piperidine-2-carboxylic acid, Lysine cyclodeaminase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | 著者 | Min, K.J, Yoon, H.J, Matsuura, A, Kim, Y.H, Lee, H.H. | | 登録日 | 2017-11-20 | | 公開日 | 2018-05-02 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.923 Å) | | 主引用文献 | Structural Basis for Recognition of L-lysine, L-ornithine, and L-2,4-diamino Butyric Acid by Lysine Cyclodeaminase.
Mol. Cells, 41, 2018
|
|
5YU3
 
 | | Structural basis for recognition of L-lysine, L-ornithine, and L-2,4-diamino butyric acid by lysine cyclodeaminase | | 分子名称: | Lysine cyclodeaminase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PROLINE, ... | | 著者 | Min, K.J, Yoon, H.J, Matsuura, A, Kim, Y.H, Lee, H.H. | | 登録日 | 2017-11-20 | | 公開日 | 2018-05-02 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Structural Basis for Recognition of L-lysine, L-ornithine, and L-2,4-diamino Butyric Acid by Lysine Cyclodeaminase.
Mol. Cells, 41, 2018
|
|
6CXX
 
 | | Horse liver E267H alcohol dehydrogenase complex with 3'-dephosphocoenzyme A | | 分子名称: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, Alcohol dehydrogenase E chain, ... | | 著者 | Plapp, B.V. | | 登録日 | 2018-04-04 | | 公開日 | 2018-04-25 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.26 Å) | | 主引用文献 | Substitutions of a buried glutamate residue hinder the conformational change in horse liver alcohol dehydrogenase and yield a surprising complex with endogenous 3'-Dephosphocoenzyme A.
Arch. Biochem. Biophys., 653, 2018
|
|
6CY3
 
 | | Horse liver E267N alcohol dehydrogenase complex with 3'-dephosphocoenzyme A | | 分子名称: | DEPHOSPHO COENZYME A, ZINC ION, alcohol dehydrogenase | | 著者 | Plapp, B.V. | | 登録日 | 2018-04-04 | | 公開日 | 2018-04-18 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Substitutions of a buried glutamate residue hinder the conformational change in horse liver alcohol dehydrogenase and yield a surprising complex with endogenous 3'-Dephosphocoenzyme A.
Arch. Biochem. Biophys., 653, 2018
|
|
4Z85
 
 | |
8X9G
 
 | | Crystal structure of CO dehydrogenase mutant in complex with BV | | 分子名称: | 1-(phenylmethyl)-4-[1-(phenylmethyl)pyridin-1-ium-4-yl]pyridin-1-ium, Carbon monoxide dehydrogenase 2, FE(4)-NI(1)-S(4) CLUSTER, ... | | 著者 | Lee, H.H, Heo, Y, Yoon, H.J, Kim, S.M, Kong, S.Y. | | 登録日 | 2023-11-30 | | 公開日 | 2024-04-17 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (3.11 Å) | | 主引用文献 | Identifying a key spot for electron mediator-interaction to tailor CO dehydrogenase's affinity.
Nat Commun, 15, 2024
|
|
8X9H
 
 | | Crystal structure of CO dehydrogenase mutant (F41C) | | 分子名称: | Carbon monoxide dehydrogenase 2, FE (III) ION, FE(4)-NI(1)-S(4) CLUSTER, ... | | 著者 | Lee, H.H, Heo, Y, Yoon, H.J, Kim, S.M, Kong, S.Y. | | 登録日 | 2023-11-30 | | 公開日 | 2024-04-17 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Identifying a key spot for electron mediator-interaction to tailor CO dehydrogenase's affinity.
Nat Commun, 15, 2024
|
|
8X9F
 
 | | Crystal structure of CO dehydrogenase mutant in complex with EV | | 分子名称: | 1,2-ETHANEDIOL, 1-ethyl-4-(1-ethylpyridin-1-ium-4-yl)pyridin-1-ium, Carbon monoxide dehydrogenase 2, ... | | 著者 | Lee, H.H, Heo, Y, Yoon, H.J, Kim, S.M, Kong, S.Y. | | 登録日 | 2023-11-30 | | 公開日 | 2024-04-17 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.48 Å) | | 主引用文献 | Identifying a key spot for electron mediator-interaction to tailor CO dehydrogenase's affinity.
Nat Commun, 15, 2024
|
|
8X9E
 
 | | Crystal structure of CO dehydrogenase mutant with increased affinity for electron mediators in low PEG concentration | | 分子名称: | 1,2-ETHANEDIOL, Carbon monoxide dehydrogenase 2, FE (III) ION, ... | | 著者 | Lee, H.H, Heo, Y, Yoon, H.J, Kim, S.M, Kong, S.Y. | | 登録日 | 2023-11-30 | | 公開日 | 2024-04-17 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Identifying a key spot for electron mediator-interaction to tailor CO dehydrogenase's affinity.
Nat Commun, 15, 2024
|
|
8X9D
 
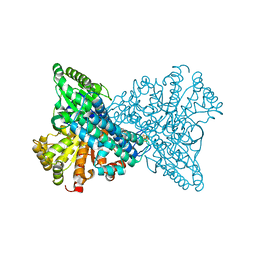 | | Crystal structure of CO dehydrogenase mutant with increased affinity for electron mediators in high PEG concentration | | 分子名称: | Carbon monoxide dehydrogenase 2, FE (III) ION, FE(4)-NI(1)-S(4) CLUSTER, ... | | 著者 | Lee, H.H, Heo, Y, Yoon, H.J, Kim, S.M, Kong, S.Y. | | 登録日 | 2023-11-30 | | 公開日 | 2024-04-17 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.11 Å) | | 主引用文献 | Identifying a key spot for electron mediator-interaction to tailor CO dehydrogenase's affinity.
Nat Commun, 15, 2024
|
|
2X29
 
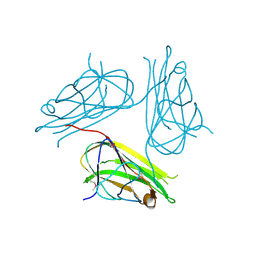 | | Crystal structure of human4-1BB ligand ectodomain | | 分子名称: | TUMOR NECROSIS FACTOR LIGAND SUPERFAMILY MEMBER 9 | | 著者 | Won, E.Y, Cho, H.S. | | 登録日 | 2010-01-12 | | 公開日 | 2010-03-23 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | The Structure of the Trimer of Human 4-1Bb Ligand is Unique Among Members of the Tumor Necrosis Factor Superfamily.
J.Biol.Chem., 285, 2010
|
|
7ERR
 
 | |
5JCK
 
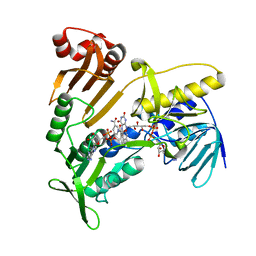 | | Structure and catalytic mechanism of monodehydroascorbate reductase, MDHAR, from Oryza sativa L. japonica | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Os09g0567300 protein | | 著者 | Park, A.K, Kim, H.W. | | 登録日 | 2016-04-15 | | 公開日 | 2016-10-12 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure and catalytic mechanism of monodehydroascorbate reductase, MDHAR, from Oryza sativa L. japonica
Sci Rep, 6
|
|
5JCN
 
 | | Structure and catalytic mechanism of monodehydroascorbate reductase, MDHAR, from Oryza sativa L. japonica | | 分子名称: | ASCORBIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | 著者 | Park, A.K, Kim, H.W. | | 登録日 | 2016-04-15 | | 公開日 | 2016-10-12 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.29 Å) | | 主引用文献 | Structure and catalytic mechanism of monodehydroascorbate reductase, MDHAR, from Oryza sativa L. japonica
Sci Rep, 6, 2016
|
|
5JCL
 
 | | Structure and catalytic mechanism of monodehydroascorbate reductase, MDHAR, from Oryza sativa L. japonica | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Os09g0567300 protein | | 著者 | Park, A.K, Kim, H.W. | | 登録日 | 2016-04-15 | | 公開日 | 2016-10-12 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structure and catalytic mechanism of monodehydroascorbate reductase, MDHAR, from Oryza sativa L. japonica
Sci Rep, 6, 2016
|
|
5JCI
 
 | | Structure and catalytic mechanism of monodehydroascorbate reductase, MDHAR, from Oryza sativa L. japonica | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, Os09g0567300 protein | | 著者 | Park, A.K, Kim, H.W. | | 登録日 | 2016-04-15 | | 公開日 | 2016-10-12 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structure and catalytic mechanism of monodehydroascorbate reductase, MDHAR, from Oryza sativa L. japonica
Sci Rep, 6, 2016
|
|
5JCM
 
 | | Structure and catalytic mechanism of monodehydroascorbate reductase, MDHAR, from Oryza sativa L. japonica | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, ISOASCORBIC ACID, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | 著者 | Park, A.K, Kim, H.W. | | 登録日 | 2016-04-15 | | 公開日 | 2016-10-12 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure and catalytic mechanism of monodehydroascorbate reductase, MDHAR, from Oryza sativa L. japonica
Sci Rep, 6, 2016
|
|
5H63
 
 | | Structure of Transferase mutant-C23S,C199S | | 分子名称: | MANGANESE (II) ION, Transferase, URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE | | 著者 | Park, J.B, Yoo, Y, Kim, J. | | 登録日 | 2016-11-10 | | 公開日 | 2017-12-20 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | Structural basis for arginine glycosylation of host substrates by bacterial effector proteins.
Nat Commun, 9, 2018
|
|
