7Z79
 
 | | Crystal structure of aminotransferase-like protein from Variovorax paradoxus | | 分子名称: | Aminotransferase, class 4, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Boyko, K.M, Matyuta, I.O, Nikolaeva, A.Y, Khrenova, M.G, Rakitina, T.V, Popov, V.O, Bezsudnova, E.Y. | | 登録日 | 2022-03-15 | | 公開日 | 2022-04-13 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | A Puzzling Protein from Variovorax paradoxus Has a PLP Fold Type IV Transaminase Structure and Binds PLP without Catalytic Lysine
Crystals, 12, 2022
|
|
7Q6B
 
 | | mRubyFT/S148I, a mutant of blue-to-red fluorescent timer in its blue state | | 分子名称: | mRubyFT S148I, a mutant of blue-to-red fluorescent timer | | 著者 | Boyko, K.M, Nikolaeva, A.Y, Vlaskina, A.V, Dorovatovskii, P.V, Khrenova, M.G, Subach, O.M, Popov, V.O, Subach, F.M. | | 登録日 | 2021-11-06 | | 公開日 | 2023-04-12 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Combined Structural and Computational Study of the mRubyFT Fluorescent Timer Locked in Its Blue Form.
Int J Mol Sci, 24, 2023
|
|
7OIN
 
 | | Crystal structure of LSSmScarlet - a genetically encoded red fluorescent protein with a large Stokes shift | | 分子名称: | LSSmScarlet - Genetically Encoded Red Fluorescent Proteins with a Large Stokes Shift, SODIUM ION, SULFATE ION | | 著者 | Boyko, K.M, Nikolaeva, A.Y, Dorovatovskii, P.V, Subach, O.M, Vlaskina, A.V, Agapova, Y.K, Ivashkina, O.I, Popov, V.O, Subach, F.V. | | 登録日 | 2021-05-12 | | 公開日 | 2022-02-16 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | LSSmScarlet, dCyRFP2s, dCyOFP2s and CRISPRed2s, Genetically Encoded Red Fluorescent Proteins with a Large Stokes Shift.
Int J Mol Sci, 22, 2021
|
|
5OEX
 
 | | Complex with iodine ion for thiocyanate dehydrogenase from Thioalkalivibrio paradoxus | | 分子名称: | 1,2-ETHANEDIOL, 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, COPPER (II) ION, ... | | 著者 | Polyakov, K.M, Tsallagov, S.I, Tikhonova, T.V, Popov, V.O. | | 登録日 | 2017-07-10 | | 公開日 | 2018-08-01 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Discovery and characterization of a novel copper containing enzyme - THIOCYANATE DEHYDROGENASE.
To Be Published
|
|
6I3Q
 
 | | The structure of thiocyanate dehydrogenase from Thioalkalivibrio paradoxus complex with acetate ions. | | 分子名称: | ACETATE ION, COPPER (II) ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Polyakov, K.M, Popov, A.N, Tikhkonova, T.V, Popov, V.O, Trofimov, A.A. | | 登録日 | 2018-11-07 | | 公開日 | 2018-11-28 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Trinuclear copper biocatalytic center forms an active site of thiocyanate dehydrogenase.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6QVM
 
 | | Undecaheme cytochrome from S-layer of Carboxydothermus ferrireducens | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose, ... | | 著者 | Osipov, E.M, Dergousova, N.I, Boyko, K.M, Tikhonova, T.V, Gavrilov, S.F, Popov, V.O. | | 登録日 | 2019-03-04 | | 公開日 | 2020-03-25 | | 最終更新日 | 2023-02-08 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Extracellular Fe(III) reductase structure reveals a modular organization enabling S-layer insertion and electron transfer to insoluble substrates
Structure, 2023
|
|
7P7X
 
 | | Crystal structure of D-amino acid transaminase from Haliscomenobacter hydrossis (holo form). | | 分子名称: | ACETATE ION, Aminotransferase class IV, PHOSPHATE ION, ... | | 著者 | Boyko, K.M, Nikolaeva, A.Y, Bakunova, A.K, Rakitina, T.V, Bezsudnova, E.Y, Popov, V.O. | | 登録日 | 2021-07-20 | | 公開日 | 2021-07-28 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The Uncommon Active Site of D-Amino Acid Transaminase from Haliscomenobacter hydrossis : Biochemical and Structural Insights into the New Enzyme.
Molecules, 26, 2021
|
|
6SJI
 
 | | The structure of thiocyanate dehydrogenase from Thioalkalivibrio paradoxus mutant with His 482 replaced by Gln | | 分子名称: | COPPER (II) ION, SULFATE ION, thiocyanate dehydrogenase | | 著者 | Polyakov, K.M, Tikhonova, T.V, Rakitina, T.V, Osipov, E, Popov, V.O. | | 登録日 | 2019-08-13 | | 公開日 | 2019-09-18 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Trinuclear copper biocatalytic center forms an active site of thiocyanate dehydrogenase.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6G50
 
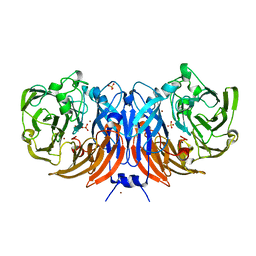 | | The structure of thiocyanate dehydrogenase from Thioalkalivibrio paradoxus as isolated. | | 分子名称: | 1,2-ETHANEDIOL, COPPER (II) ION, SULFATE ION, ... | | 著者 | Polyakov, K.M, Tsallagov, S.I, Tikhkonova, T.V, Popov, V.O. | | 登録日 | 2018-03-28 | | 公開日 | 2019-04-10 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Trinuclear copper biocatalytic center forms an active site of thiocyanate dehydrogenase.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6UWE
 
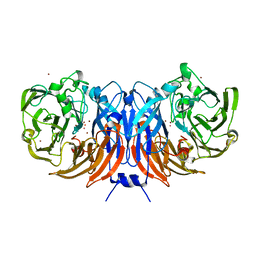 | | Crystal structure of recombinant thiocyanate dehydrogenase from Thioalkalivibrio paradoxus saturated with copper | | 分子名称: | COPPER (II) ION, UNKNOWN ATOM OR ION, thiocyanate dehydrogenase | | 著者 | Shabalin, I.G, Osipov, E, Tikhonova, T.V, Rakitina, T.V, Boyko, K.M, Popov, V.O. | | 登録日 | 2019-11-05 | | 公開日 | 2019-11-27 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Trinuclear copper biocatalytic center forms an active site of thiocyanate dehydrogenase.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8ONJ
 
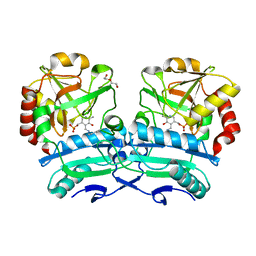 | | Crystal structure of D-amino acid aminotransferase from Aminobacterium colombiense point mutant R88L | | 分子名称: | Aminotransferase class IV, DI(HYDROXYETHYL)ETHER, PYRIDOXAL-5'-PHOSPHATE | | 著者 | Matyuta, I.O, Boyko, K.M, Minyaev, M.E, Shilova, S.A, Bezsudnova, E.Y, Popov, V.O. | | 登録日 | 2023-04-03 | | 公開日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | In search for structural targets for engineering d-amino acid transaminase: modulation of pH optimum and substrate specificity.
Biochem.J., 480, 2023
|
|
8ONL
 
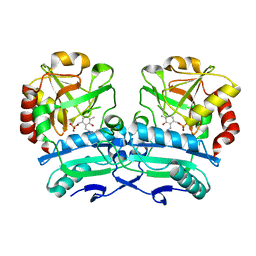 | | Crystal structure of D-amino acid aminotransferase from Aminobacterium colombiense point mutant E113A | | 分子名称: | Aminotransferase class IV, PYRIDOXAL-5'-PHOSPHATE | | 著者 | Matyuta, I.O, Boyko, K.M, Minyaev, M.E, Shilova, S.A, Bezsudnova, E.Y, Popov, V.O. | | 登録日 | 2023-04-03 | | 公開日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | In search for structural targets for engineering d-amino acid transaminase: modulation of pH optimum and substrate specificity.
Biochem.J., 480, 2023
|
|
8ONN
 
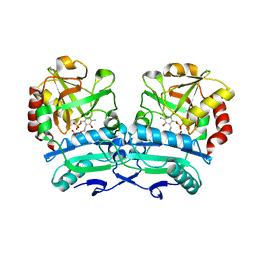 | | Crystal structure of D-amino acid aminotransferase from Aminobacterium colombiense point mutant E113A complexed with 3-aminooxypropionic acid | | 分子名称: | 3-[(~{E})-[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylideneamino]oxypropanoic acid, Aminotransferase class IV | | 著者 | Matyuta, I.O, Boyko, K.M, Minyaev, M.E, Shilova, S.A, Bezsudnova, E.Y, Popov, V.O. | | 登録日 | 2023-04-03 | | 公開日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | In search for structural targets for engineering d-amino acid transaminase: modulation of pH optimum and substrate specificity.
Biochem.J., 480, 2023
|
|
8ONM
 
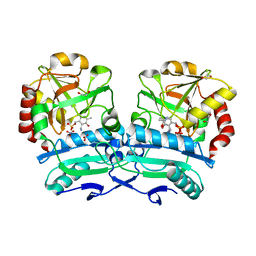 | | Crystal structure of D-amino acid aminotransferase from Aminobacterium colombiense point mutant E113A complexed with D-glutamate | | 分子名称: | (~{Z})-2-[[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylamino]pent-2-enedioic acid, 1,2-ETHANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, ... | | 著者 | Matyuta, I.O, Boyko, K.M, Minyaev, M.E, Shilova, S.A, Bezsudnova, E.Y, Popov, V.O. | | 登録日 | 2023-04-03 | | 公開日 | 2024-04-10 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Probing of the structural and catalytic roles of the residues in the active site of transaminase from Aminobacterium colombiense
To Be Published
|
|
8PYH
 
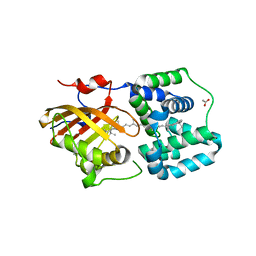 | | Crystal structure of the Orange Carotenoid Protein 2 (OCP2) from Crinalium epipsammum PCC 9333 | | 分子名称: | ACETATE ION, Orange carotenoid-binding protein, beta,beta-caroten-4-one | | 著者 | Boyko, K.M, Varfolomeeva, L.A, Slonimskiy, Y.B, Maksimov, E.G, Popov, V.O, Sluchanko, N.N. | | 登録日 | 2023-07-25 | | 公開日 | 2024-01-03 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural framework for the understanding spectroscopic and functional signatures of the cyanobacterial Orange Carotenoid Protein families.
Int.J.Biol.Macromol., 254, 2024
|
|
8PZK
 
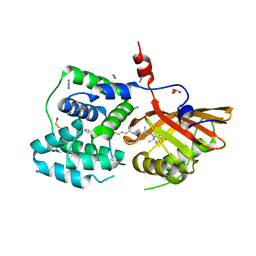 | | Crystal structure of the Orange Carotenoid Protein 2 (OCP2) from Gloeocapsa sp. PCC 7428 | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, AZIDE ION, GLYCEROL, ... | | 著者 | Boyko, K.M, Varfolomeeva, L.A, Slonimskiy, Y.B, Maksimov, E.G, Popov, V.O, Sluchanko, N.N. | | 登録日 | 2023-07-27 | | 公開日 | 2024-01-03 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural framework for the understanding spectroscopic and functional signatures of the cyanobacterial Orange Carotenoid Protein families.
Int.J.Biol.Macromol., 254, 2024
|
|
8AHR
 
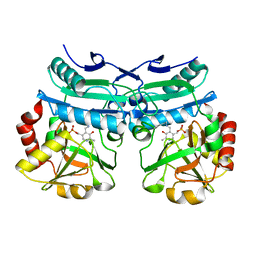 | | Crystal structure of D-amino acid aminotransferase from Aminobacterium colombiense in holo form with PLP | | 分子名称: | Aminotransferase class IV, PYRIDOXAL-5'-PHOSPHATE | | 著者 | Matyuta, I.O, Boyko, K.M, Nikolaeva, A.Y, Shilova, S.A, Rakitina, T.V, Popov, V.O, Bezsudnova, E.Y. | | 登録日 | 2022-07-22 | | 公開日 | 2022-08-03 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | To the Understanding of Catalysis by D-Amino Acid Transaminases: A Case Study of the Enzyme from Aminobacterium colombiense.
Molecules, 28, 2023
|
|
8AYJ
 
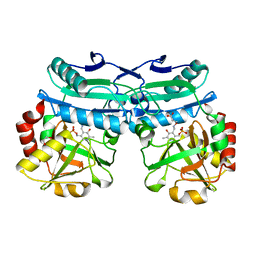 | | Crystal structure of D-amino acid aminotransferase from Aminobacterium colombiens complexed with 3-aminooxypropionic acid | | 分子名称: | 1,2-ETHANEDIOL, 3-[(~{E})-[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylideneamino]oxypropanoic acid, Aminotransferase class IV, ... | | 著者 | Matyuta, I.O, Boyko, K.M, Nikolaeva, A.Y, Shilova, S.A, Rakitina, T.V, Popov, V.O, Bezsudnova, E.Y. | | 登録日 | 2022-09-02 | | 公開日 | 2022-11-16 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | In search for structural targets for engineering d-amino acid transaminase: modulation of pH optimum and substrate specificity.
Biochem.J., 480, 2023
|
|
8AYK
 
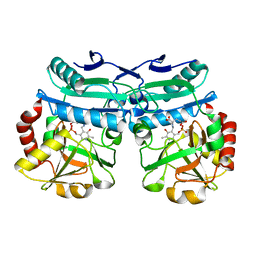 | | Crystal structure of D-amino acid aminotrensferase from Aminobacterium colombiense complexed with D-glutamate | | 分子名称: | (~{Z})-2-[[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylamino]pent-2-enedioic acid, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Aminotransferase class IV | | 著者 | Matyuta, I.O, Boyko, K.M, Nikolaeva, A.Y, Shilova, S.A, Rakitina, T.V, Minyaev, M.E, Popov, V.O, Bezsudnova, E.Y. | | 登録日 | 2022-09-02 | | 公開日 | 2022-11-16 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | To the Understanding of Catalysis by D-Amino Acid Transaminases: A Case Study of the Enzyme from Aminobacterium colombiense.
Molecules, 28, 2023
|
|
8PNY
 
 | | Crystal structure of D-amino acid aminotransferase from Blastococcus saxobsidens complexed with phenylhydrazine and in its apo form | | 分子名称: | Branched-chain amino acid aminotransferase/4-amino-4-deoxychorismate lyase, [6-methyl-5-oxidanyl-4-[(2-phenylhydrazinyl)methyl]pyridin-3-yl]methyl dihydrogen phosphate | | 著者 | Matyuta, I.O, Boyko, K.M, Nikolaeva, A.Y, Shilova, S.A, Popov, V.O. | | 登録日 | 2023-07-03 | | 公開日 | 2023-10-25 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Expanded Substrate Specificity in D-Amino Acid Transaminases: A Case Study of Transaminase from Blastococcus saxobsidens.
Int J Mol Sci, 24, 2023
|
|
8PNW
 
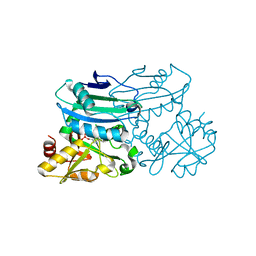 | | Crystal structure of D-amino acid aminotransferase from Blastococcus saxobsidens in holo form with PLP | | 分子名称: | Branched-chain amino acid aminotransferase/4-amino-4-deoxychorismate lyase, CHLORIDE ION, PYRIDOXAL-5'-PHOSPHATE | | 著者 | Matyuta, I.O, Boyko, K.M, Nikolaeva, A.Y, Shilova, S.A, Popov, V.O. | | 登録日 | 2023-07-03 | | 公開日 | 2023-10-25 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Expanded Substrate Specificity in D-Amino Acid Transaminases: A Case Study of Transaminase from Blastococcus saxobsidens.
Int J Mol Sci, 24, 2023
|
|
