3FY4
 
 | | (6-4) Photolyase Crystal Structure | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-4 photolyase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | 著者 | Hitomi, K, Arvai, A.S, Tainer, J.A, Getzoff, E.D. | | 登録日 | 2009-01-21 | | 公開日 | 2009-04-28 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Functional motifs in the (6-4) photolyase crystal structure make a comparative framework for DNA repair photolyases and clock cryptochromes.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
1WWJ
 
 | | crystal structure of KaiB from Synechocystis sp. | | 分子名称: | Circadian clock protein kaiB, D-MALATE, IMIDAZOLE, ... | | 著者 | Hitomi, K, Oyama, T, Han, S, Arvai, A.S, Tainer, J.A, Getzoff, E.D. | | 登録日 | 2005-01-06 | | 公開日 | 2005-02-15 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Tetrameric architecture of the circadian clock protein KaiB. A novel interface for intermolecular interactions and its impact on the circadian rhythm.
J.Biol.Chem., 280, 2005
|
|
4EEU
 
 | | Crystal structure of phiLOV2.1 | | 分子名称: | FLAVIN MONONUCLEOTIDE, Phototropin-2 | | 著者 | Hitomi, K, Christie, J.M, Arvai, A.S, Hartfield, K.A, Pratt, A.J, Tainer, J.A, Getzoff, E.D. | | 登録日 | 2012-03-28 | | 公開日 | 2012-05-16 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.4068 Å) | | 主引用文献 | Structural Tuning of the Fluorescent Protein iLOV for Improved Photostability.
J.Biol.Chem., 287, 2012
|
|
3UMV
 
 | | Eukaryotic Class II CPD photolyase structure reveals a basis for improved UV-tolerance in plants | | 分子名称: | 1,2-ETHANEDIOL, Deoxyribodipyrimidine photo-lyase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | 著者 | Arvai, A.S, Hitomi, K, Getzoff, E.D, Tainer, J.A. | | 登録日 | 2011-11-14 | | 公開日 | 2011-12-21 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.705 Å) | | 主引用文献 | Eukaryotic Class II Cyclobutane Pyrimidine Dimer Photolyase Structure Reveals Basis for Improved Ultraviolet Tolerance in Plants.
J.Biol.Chem., 287, 2012
|
|
4EER
 
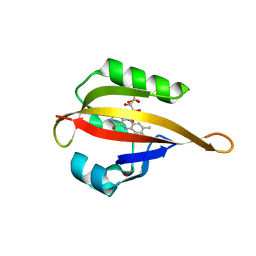 | | Crystal structure of LOV2 domain of Arabidopsis thaliana phototropin 2 C426A mutant | | 分子名称: | FLAVIN MONONUCLEOTIDE, Phototropin-2 | | 著者 | Hitomi, K, Christie, J.M, Arvai, A.S, Hartfield, K.A, Pratt, A.J, Tainer, J.A, Getzoff, E.D. | | 登録日 | 2012-03-28 | | 公開日 | 2012-05-16 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.753 Å) | | 主引用文献 | Structural Tuning of the Fluorescent Protein iLOV for Improved Photostability.
J.Biol.Chem., 287, 2012
|
|
4EES
 
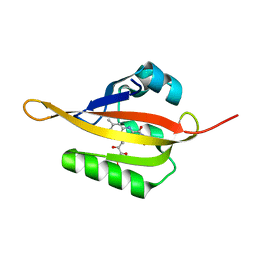 | | Crystal structure of iLOV | | 分子名称: | FLAVIN MONONUCLEOTIDE, Phototropin-2 | | 著者 | Hitomi, K, Christie, J.M, Arvai, A.S, Hartfield, K.A, Pratt, A.J, Tainer, J.A, Getzoff, E.D. | | 登録日 | 2012-03-28 | | 公開日 | 2012-05-16 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.805 Å) | | 主引用文献 | Structural Tuning of the Fluorescent Protein iLOV for Improved Photostability.
J.Biol.Chem., 287, 2012
|
|
4EET
 
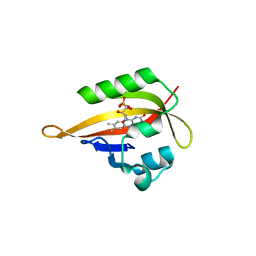 | | Crystal structure of iLOV | | 分子名称: | FLAVIN MONONUCLEOTIDE, Phototropin-2 | | 著者 | Hitomi, K, Christie, J.M, Arvai, A.S, Hartfield, K.A, Pratt, A.J, Tainer, J.A, Getzoff, E.D. | | 登録日 | 2012-03-28 | | 公開日 | 2012-05-16 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Structural Tuning of the Fluorescent Protein iLOV for Improved Photostability.
J.Biol.Chem., 287, 2012
|
|
4EEP
 
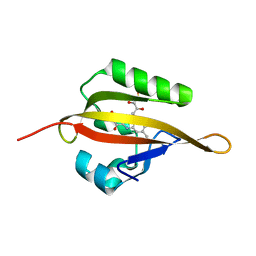 | | Crystal structure of LOV2 domain of Arabidopsis thaliana phototropin 2 | | 分子名称: | FLAVIN MONONUCLEOTIDE, Phototropin-2 | | 著者 | Hitomi, K, Christie, J.M, Arvai, A.S, Hartfield, K.A, Pratt, A.J, Tainer, J.A, Getzoff, E.D. | | 登録日 | 2012-03-28 | | 公開日 | 2012-05-16 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural Tuning of the Fluorescent Protein iLOV for Improved Photostability.
J.Biol.Chem., 287, 2012
|
|
1WEG
 
 | | Catalytic Domain Of Muty From Escherichia Coli K142A Mutant | | 分子名称: | 1,2-ETHANEDIOL, A/G-specific adenine glycosylase, IMIDAZOLE, ... | | 著者 | Hitomi, K, Arvai, A.S, Tainer, J.A. | | 登録日 | 2004-05-25 | | 公開日 | 2004-09-21 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Reaction intermediates in the catalytic mechanism of Escherichia coli MutY DNA glycosylase
J.Biol.Chem., 279, 2004
|
|
1WEF
 
 | |
1WEI
 
 | | Catalytic Domain Of Muty From Escherichia Coli K20A Mutant Complexed To Adenine | | 分子名称: | 1,2-ETHANEDIOL, A/G-specific adenine glycosylase, ADENINE, ... | | 著者 | Hitomi, K, Arvai, A.S, Tainer, J.A. | | 登録日 | 2004-05-25 | | 公開日 | 2004-09-21 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Reaction intermediates in the catalytic mechanism of Escherichia coli MutY DNA glycosylase
J.Biol.Chem., 279, 2004
|
|
3K3K
 
 | | Crystal structure of dimeric abscisic acid (ABA) receptor pyrabactin resistance 1 (PYR1) with ABA-bound closed-lid and ABA-free open-lid subunits | | 分子名称: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Abscisic acid receptor PYR1 | | 著者 | Arvai, A.S, Hitomi, K, Getzoff, E.D. | | 登録日 | 2009-10-02 | | 公開日 | 2009-11-17 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural mechanism of abscisic acid binding and signaling by dimeric PYR1.
Science, 326, 2009
|
|
4D9S
 
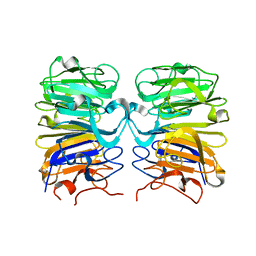 | | Crystal structure of Arabidopsis thaliana UVR8 (UV Resistance locus 8) | | 分子名称: | UVB-resistance protein UVR8 | | 著者 | Arvai, A.S, Christie, J.M, Pratt, A.J, Hitomi, K, Getzoff, E.D. | | 登録日 | 2012-01-11 | | 公開日 | 2012-04-04 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.701 Å) | | 主引用文献 | Plant UVR8 Photoreceptor Senses UV-B by Tryptophan-Mediated Disruption of Cross-Dimer Salt Bridges.
Science, 335, 2012
|
|
1NP7
 
 | | Crystal Structure Analysis of Synechocystis sp. PCC6803 cryptochrome | | 分子名称: | DNA photolyase, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | 著者 | Brudler, R, Hitomi, K, Daiyasu, H, Toh, H, Kucho, K, Ishiura, M, Kanehisa, M, Roberts, V.A, Todo, T, Tainer, J.A, Getzoff, E.D. | | 登録日 | 2003-01-17 | | 公開日 | 2003-01-28 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Identification of a new cryptochrome class: structure, function, and evolution
Mol.Cell, 11, 2003
|
|
4IEM
 
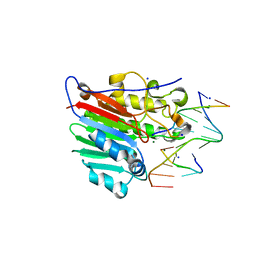 | | Human apurinic/apyrimidinic endonuclease (APE1) with product DNA and Mg2+ | | 分子名称: | DNA (5'-D(*CP*GP*AP*TP*CP*GP*GP*TP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*AP*C)-3'), DNA (5'-D(P*(3DR)P*GP*AP*TP*CP*G)-3'), ... | | 著者 | Tsutakawa, S.E, Mol, C.D, Arvai, A.S, Tainer, J.A. | | 登録日 | 2012-12-13 | | 公開日 | 2013-01-23 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.3936 Å) | | 主引用文献 | Conserved Structural Chemistry for Incision Activity in Structurally Non-homologous Apurinic/Apyrimidinic Endonuclease APE1 and Endonuclease IV DNA Repair Enzymes.
J.Biol.Chem., 288, 2013
|
|
4HNO
 
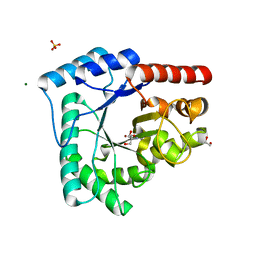 | | High resolution crystal structure of DNA Apurinic/apyrimidinic (AP) endonuclease IV Nfo from Thermatoga maritima | | 分子名称: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MAGNESIUM ION, ... | | 著者 | Shin, D.S, Hosfield, D.J, Arvai, A.S, Tsutakawa, S.E, Tainer, J.A. | | 登録日 | 2012-10-20 | | 公開日 | 2013-01-23 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (0.9194 Å) | | 主引用文献 | Conserved Structural Chemistry for Incision Activity in Structurally Non-homologous Apurinic/Apyrimidinic Endonuclease APE1 and Endonuclease IV DNA Repair Enzymes.
J.Biol.Chem., 288, 2013
|
|
