7OC8
 
 | | Trichoderma reesei Cel7A E212Q mutant in complex with pNPL | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COBALT (II) ION, Exoglucanase 1, ... | | 著者 | Haataja, T, Sandgren, M, Stahlberg, J. | | 登録日 | 2021-04-26 | | 公開日 | 2022-03-09 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Enzyme kinetics by GH7 cellobiohydrolases on chromogenic substrates is dictated by non-productive binding: insights from crystal structures and MD simulation.
Febs J., 290, 2023
|
|
7NYT
 
 | | Trichoderma reesei Cel7A E212Q mutant in complex with lactose. | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COBALT (II) ION, Exoglucanase 1, ... | | 著者 | Haataja, T, Sandgren, M, Stahlberg, J. | | 登録日 | 2021-03-23 | | 公開日 | 2022-03-09 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.09 Å) | | 主引用文献 | Enzyme kinetics by GH7 cellobiohydrolases on chromogenic substrates is dictated by non-productive binding: insights from crystal structures and MD simulation.
Febs J., 290, 2023
|
|
8POF
 
 | | The crystal structure of RsSymEG1 reveals a unique form of smaller GH7 endoglucanases alongside GH7 cellobiohydrolases in protist symbionts of termites | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Putative glycosyl hydrolase family7, SODIUM ION | | 著者 | Haataja, T, Sandgren, M, Hansson, H, Stahlberg, J. | | 登録日 | 2023-07-04 | | 公開日 | 2023-12-27 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | The crystal structure of RsSymEG1 reveals a unique form of smaller GH7 endoglucanases alongside GH7 cellobiohydrolases in protist symbionts of termites.
Febs J., 291, 2024
|
|
4V0Z
 
 | |
4UWT
 
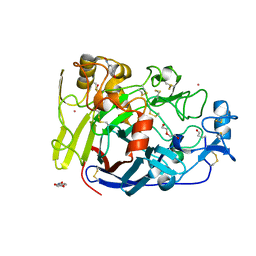 | | Hypocrea jecorina Cel7A E212Q mutant in complex with p-nitrophenyl cellobioside | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CELLOBIOHYDROLASE CEL7A, COBALT (II) ION, ... | | 著者 | Nutt, A, Momeni, M.H, Johansson, G, Stahlberg, J. | | 登録日 | 2014-08-14 | | 公開日 | 2015-08-26 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Enzyme kinetics by GH7 cellobiohydrolases on chromogenic substrates is dictated by non-productive binding: insights from crystal structures and MD simulation.
Febs J., 2022
|
|
