6A4T
 
 | | Crystal structure of Peptidase E from Deinococcus radiodurans R1 | | 分子名称: | Peptidase E | | 著者 | Yadav, P, Goyal, V.G, Kumar, A, Gokhale, S.M, Makde, R.D. | | 登録日 | 2018-06-20 | | 公開日 | 2019-06-26 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Catalytic triad heterogeneity in S51 peptidase family: Structural basis for functional variability.
Proteins, 87, 2019
|
|
6A4R
 
 | | Crystal structure of aspartate bound peptidase E from Salmonella enterica | | 分子名称: | ASPARTIC ACID, Peptidase E | | 著者 | Yadav, P, Chandravanshi, K, Goyal, V.D, Singh, R, Kumar, A, Gokhale, S.M, Makde, R.D. | | 登録日 | 2018-06-20 | | 公開日 | 2018-10-24 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.828 Å) | | 主引用文献 | Structure of Asp-bound peptidase E from Salmonella enterica: Active site at dimer interface illuminates Asp recognition.
FEBS Lett., 592, 2018
|
|
6A4S
 
 | | Crystal structure of peptidase E with ordered active site loop from Salmonella enterica | | 分子名称: | Peptidase E | | 著者 | Yadav, P, Chandravanshi, K, Goyal, V.D, Singh, R, Kumar, A, Gokhale, S.M, Makde, R.D. | | 登録日 | 2018-06-20 | | 公開日 | 2018-10-31 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure of Asp-bound peptidase E from Salmonella enterica: Active site at dimer interface illuminates Asp recognition.
FEBS Lett., 592, 2018
|
|
6IKG
 
 | | Crystal structure of substrate-bound S9 peptidase (S514A mutant) from Deinococcus radiodurans | | 分子名称: | Acyl-peptide hydrolase, putative, GLYCEROL, ... | | 著者 | Yadav, P, Kumar, A, Goyal, V.D, Makde, R.D. | | 登録日 | 2018-10-16 | | 公開日 | 2018-11-14 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Carboxypeptidase in prolyl oligopeptidase family: Unique enzyme activation and substrate-screening mechanisms.
J.Biol.Chem., 294, 2019
|
|
6IRU
 
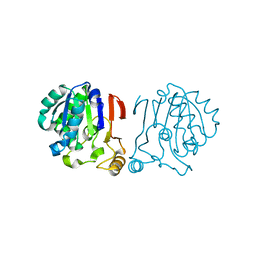 | |
6IGR
 
 | | Crystal structure of S9 peptidase (S514A mutant in inactive state) from Deinococcus radiodurans R1 | | 分子名称: | Acyl-peptide hydrolase, putative, GLYCEROL | | 著者 | Yadav, P, Gaur, N.K, Goyal, V.D, Kumar, A, Makde, R.D. | | 登録日 | 2018-09-25 | | 公開日 | 2018-11-14 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Carboxypeptidase in prolyl oligopeptidase family: Unique enzyme activation and substrate-screening mechanisms.
J.Biol.Chem., 294, 2019
|
|
6IGQ
 
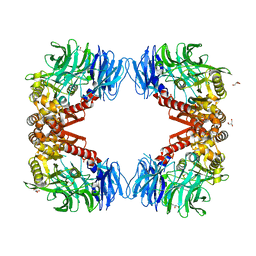 | | Crystal structure of inactive state of S9 peptidase from Deinococcus radiodurans R1 (PMSF treated) | | 分子名称: | Acyl-peptide hydrolase, putative, GLYCEROL, ... | | 著者 | Yadav, P, Goyal, V.D, Kumar, A, Makde, R.D. | | 登録日 | 2018-09-25 | | 公開日 | 2018-11-14 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Carboxypeptidase in prolyl oligopeptidase family: Unique enzyme activation and substrate-screening mechanisms.
J.Biol.Chem., 294, 2019
|
|
6IGP
 
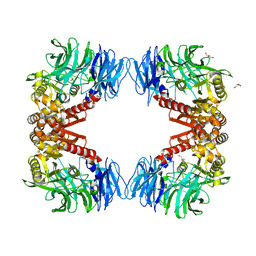 | | Crystal structure of S9 peptidase (inactive state)from Deinococcus radiodurans R1 in P212121 | | 分子名称: | Acyl-peptide hydrolase, putative, GLYCEROL | | 著者 | Yadav, P, Goyal, V.D, Kumar, A, Makde, R.D. | | 登録日 | 2018-09-25 | | 公開日 | 2018-11-14 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Carboxypeptidase in prolyl oligopeptidase family: Unique enzyme activation and substrate-screening mechanisms.
J.Biol.Chem., 294, 2019
|
|
5YZO
 
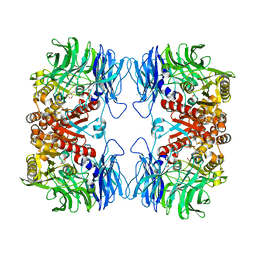 | | Crystal structure of S9 peptidase mutant (S514A) from Deinococcus radiodurans R1 | | 分子名称: | Acyl-peptide hydrolase, putative, DIMETHYL SULFOXIDE, ... | | 著者 | Yadav, P, Jamdar, S.N, Kumar, A, Ghosh, B, Makde, R.D. | | 登録日 | 2017-12-15 | | 公開日 | 2018-11-14 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Carboxypeptidase in prolyl oligopeptidase family: Unique enzyme activation and substrate-screening mechanisms.
J.Biol.Chem., 294, 2019
|
|
5YZN
 
 | | Crystal structure of S9 peptidase (active form) from Deinococcus radiodurans R1 | | 分子名称: | Acyl-peptide hydrolase, putative | | 著者 | Yadav, P, Jamdar, S.N, Kumar, A, Ghosh, B, Makde, R.D. | | 登録日 | 2017-12-15 | | 公開日 | 2018-11-14 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Carboxypeptidase in prolyl oligopeptidase family: Unique enzyme activation and substrate-screening mechanisms.
J.Biol.Chem., 294, 2019
|
|
5YZM
 
 | | Crystal structure of S9 peptidase (inactive form) from Deinococcus radiodurans R1 | | 分子名称: | ACETATE ION, Acyl-peptide hydrolase, putative | | 著者 | Yadav, P, Jamdar, S.N, Kumar, A, Ghosh, B, Makde, R.D. | | 登録日 | 2017-12-15 | | 公開日 | 2018-11-14 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Carboxypeptidase in prolyl oligopeptidase family: Unique enzyme activation and substrate-screening mechanisms.
J.Biol.Chem., 294, 2019
|
|
