6GOC
 
 | | Methylesterase BT1017 | | 分子名称: | DUF3826 domain-containing protein, ZINC ION | | 著者 | Basle, A, Ndeh, D, Gilbert, H. | | 登録日 | 2018-06-01 | | 公開日 | 2019-06-19 | | 最終更新日 | 2020-04-22 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Characterisation of a methylesterases essential for pectin rhamnogalacturonan II metabolism from the gut bacterium Bacteroides thetaiotaomicron
to be published
|
|
4RVA
 
 | | A triple mutant in the omega-loop of TEM-1 beta-lactamase changes the substrate profile via a large conformational change and an altered general base for deacylation | | 分子名称: | BICARBONATE ION, Beta-lactamase TEM | | 著者 | Stojanoski, V, Chow, D.-C, Hu, L, Sankaran, B, Gilbert, H, Prasad, B.V.V, Palzkill, T. | | 登録日 | 2014-11-25 | | 公開日 | 2015-03-04 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.4397 Å) | | 主引用文献 | A Triple Mutant in the Omega-loop of TEM-1 beta-Lactamase Changes the Substrate Profile via a Large Conformational Change and an Altered General Base for Catalysis.
J.Biol.Chem., 290, 2015
|
|
4RX3
 
 | | A triple mutant in the omega-loop of TEM-1 beta-lactamase changes the substrate profile via a large conformational change and an altered general base for catalysis | | 分子名称: | Beta-lactamase TEM, CITRATE ANION | | 著者 | Stojanoski, V, Chow, D, Hu, L, Sankaran, B, Gilbert, H, Prasad, B.V.V, Palzkill, T. | | 登録日 | 2014-12-08 | | 公開日 | 2015-03-04 | | 最終更新日 | 2017-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.39 Å) | | 主引用文献 | A Triple Mutant in the Omega-loop of TEM-1 beta-Lactamase Changes the Substrate Profile via a Large Conformational Change and an Altered General Base for Catalysis.
J.Biol.Chem., 290, 2015
|
|
4RX2
 
 | | A triple mutant in the omega-loop of TEM-1 beta-lactamase changes the substrate profile via a large conformational change and an altered general base for catalysis | | 分子名称: | Beta-lactamase TEM, SULFATE ION | | 著者 | Stojanoski, V, Chow, D, Hu, L, Sankaran, B, Gilbert, H, Prasad, B.V.V, Palzkill, T. | | 登録日 | 2014-12-08 | | 公開日 | 2015-03-04 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.315 Å) | | 主引用文献 | A Triple Mutant in the Omega-loop of TEM-1 beta-Lactamase Changes the Substrate Profile via a Large Conformational Change and an Altered General Base for Catalysis.
J.Biol.Chem., 290, 2015
|
|
6EON
 
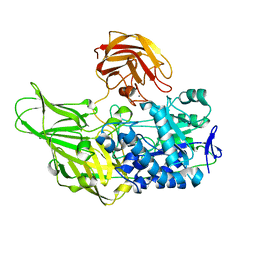 | | Galactanase BT0290 | | 分子名称: | Beta-galactosidase, CALCIUM ION, alpha-D-galactopyranose | | 著者 | Basle, A, Munoz, J, Gilbert, H. | | 登録日 | 2017-10-10 | | 公開日 | 2017-11-29 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | A surface endogalactanase in Bacteroides thetaiotaomicron confers keystone status for arabinogalactan degradation.
Nat Microbiol, 3, 2018
|
|
5NGL
 
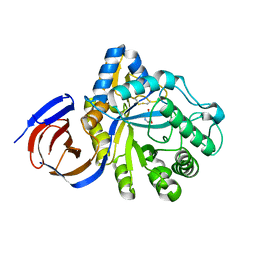 | | The endo-beta1,6-glucanase BT3312 | | 分子名称: | 1-DEOXYNOJIRIMYCIN, Glucosylceramidase, SODIUM ION, ... | | 著者 | Basle, A, Temple, M, Cuskin, F, Lowe, E, Gilbert, H. | | 登録日 | 2017-03-17 | | 公開日 | 2017-05-10 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | A Bacteroidetes locus dedicated to fungal 1,6-beta-glucan degradation: Unique substrate conformation drives specificity of the key endo-1,6-beta-glucanase.
J. Biol. Chem., 292, 2017
|
|
5NGK
 
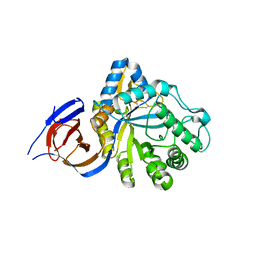 | | The endo-beta1,6-glucanase BT3312 | | 分子名称: | Glucosylceramidase | | 著者 | Basle, A, Temple, M, Cuskin, F, Lowe, E, Gilbert, H. | | 登録日 | 2017-03-17 | | 公開日 | 2017-05-10 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | A Bacteroidetes locus dedicated to fungal 1,6-beta-glucan degradation: Unique substrate conformation drives specificity of the key endo-1,6-beta-glucanase.
J. Biol. Chem., 292, 2017
|
|
5LA1
 
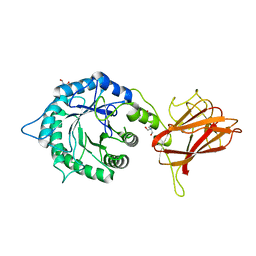 | | The mechanism by which arabinoxylanases can recognise highly decorated xylans | | 分子名称: | CALCIUM ION, Carbohydrate binding family 6, TRIS-HYDROXYMETHYL-METHYL-AMMONIUM, ... | | 著者 | Basle, A, Labourel, A, Cuskin, F, Jackson, A, Crouch, L, Rogowski, A, Gilbert, H. | | 登録日 | 2016-06-13 | | 公開日 | 2016-08-31 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | The Mechanism by Which Arabinoxylanases Can Recognize Highly Decorated Xylans.
J.Biol.Chem., 291, 2016
|
|
5LA2
 
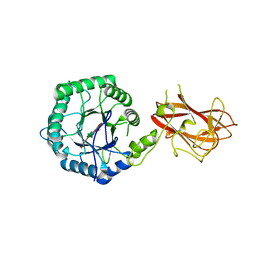 | | The mechanism by which arabinoxylanases can recognise highly decorated xylans | | 分子名称: | CALCIUM ION, Carbohydrate binding family 6, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-[alpha-L-arabinofuranose-(1-3)]alpha-D-xylopyranose, ... | | 著者 | Basle, A, Labourel, A, Cuskin, F, Jackson, A, Crouch, L, Rogowski, A, Gilbert, H. | | 登録日 | 2016-06-13 | | 公開日 | 2016-08-31 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | The Mechanism by Which Arabinoxylanases Can Recognize Highly Decorated Xylans.
J.Biol.Chem., 291, 2016
|
|
1GWK
 
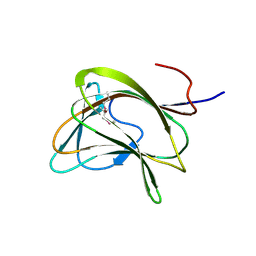 | | Carbohydrate binding module family29 | | 分子名称: | NON-CATALYTIC PROTEIN 1 | | 著者 | Charnock, S.J, Nurizzo, D, Davies, G.J. | | 登録日 | 2002-03-19 | | 公開日 | 2003-03-20 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.34 Å) | | 主引用文献 | Promiscuity in Ligand-Binding: The Three-Dimensional Structure of a Piromyces Carbohydrate-Binding Module,Cbm29-2,in Complex with Cello- and Mannohexaose
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1GWL
 
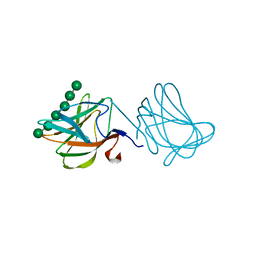 | | Carbohydrate binding module family29 complexed with mannohexaose | | 分子名称: | NON-CATALYTIC PROTEIN 1, beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose | | 著者 | Charnock, S.J, Nurizzo, D, Davies, G.J. | | 登録日 | 2002-03-19 | | 公開日 | 2003-03-20 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.51 Å) | | 主引用文献 | Promiscuity in Ligand-Binding: The Three-Dimensional Structure of a Piromyces Carbohydrate-Binding Module,Cbm29-2,in Complex with Cello- and Mannohexaose
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1GWM
 
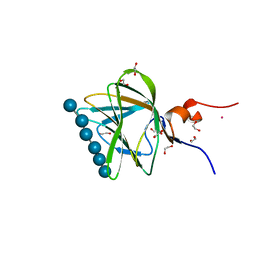 | | Carbohydrate binding module family29 complexed with glucohexaose | | 分子名称: | 1,2-ETHANEDIOL, COBALT (II) ION, NON-CATALYTIC PROTEIN 1, ... | | 著者 | Charnock, S.J, Nurizzo, D, Davies, G.J. | | 登録日 | 2002-03-19 | | 公開日 | 2003-03-20 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.15 Å) | | 主引用文献 | Promiscuity in Ligand-Binding: The Three-Dimensional Structure of a Piromyces Carbohydrate-Binding Module,Cbm29-2,in Complex with Cello- and Mannohexaose
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
6HZF
 
 | | BP0997, GH138 enzyme targeting pectin rhamnogalacturonan II | | 分子名称: | BPa0997, SERINE, SODIUM ION | | 著者 | Basle, A, Cartmell, A, Labourel, A, Gilbert, H. | | 登録日 | 2018-10-23 | | 公開日 | 2019-03-20 | | 最終更新日 | 2019-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structural and functional analyses of glycoside hydrolase 138 enzymes targeting chain A galacturonic acid in the complex pectin rhamnogalacturonan II.
J.Biol.Chem., 294, 2019
|
|
6HZE
 
 | | BP0997, GH138 enzyme targeting pectin rhamnogalacturonan II | | 分子名称: | BPa0997, SODIUM ION, beta-D-galactopyranuronic acid | | 著者 | Basle, A, Cartmell, A, Labourel, A, Gilbert, H. | | 登録日 | 2018-10-23 | | 公開日 | 2019-03-20 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structural and functional analyses of glycoside hydrolase 138 enzymes targeting chain A galacturonic acid in the complex pectin rhamnogalacturonan II.
J.Biol.Chem., 294, 2019
|
|
6HZG
 
 | | BP0997, GH138 enzyme targeting pectin rhamnogalacturonan II | | 分子名称: | BPa0997 N-ter E361S, CHLORIDE ION, SODIUM ION, ... | | 著者 | Basle, A, Cartmell, A, Labourel, A, Gilbert, H. | | 登録日 | 2018-10-23 | | 公開日 | 2019-03-20 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structural and functional analyses of glycoside hydrolase 138 enzymes targeting chain A galacturonic acid in the complex pectin rhamnogalacturonan II.
J.Biol.Chem., 294, 2019
|
|
