6C29
 
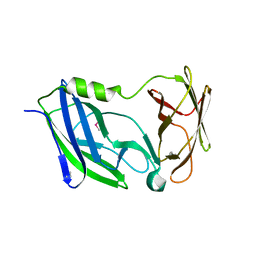 | | Crystal structure of the N-terminal periplasmic domain of ScsB from Proteus mirabilis | | 分子名称: | Putative metal resistance protein | | 著者 | Furlong, E.J, Choudhury, H.G, Kurth, F, Martin, J.L. | | 登録日 | 2018-01-07 | | 公開日 | 2018-03-07 | | 最終更新日 | 2020-01-01 | | 実験手法 | X-RAY DIFFRACTION (1.538 Å) | | 主引用文献 | Disulfide isomerase activity of the dynamic, trimericProteus mirabilisScsC protein is primed by the tandem immunoglobulin-fold domain of ScsB.
J. Biol. Chem., 293, 2018
|
|
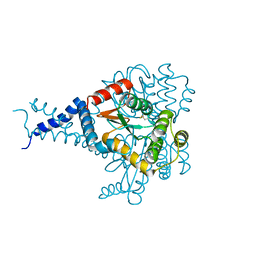
6MHH
 
 | |
5IDR
 
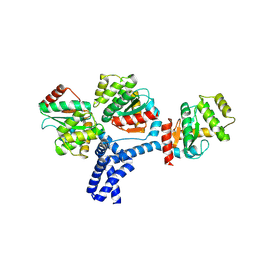 | |
4XVW
 
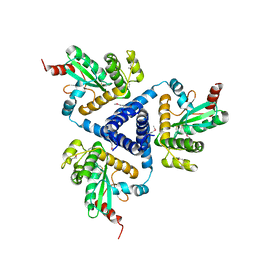 | |
6NEN
 
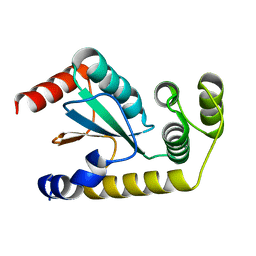 | | Catalytic domain of Proteus mirabilis ScsC | | 分子名称: | Copper resistance protein | | 著者 | Kurth, F, Furlong, E.J, Premkumar, L, Martin, J.L. | | 登録日 | 2018-12-17 | | 公開日 | 2019-03-06 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.151 Å) | | 主引用文献 | Engineered variants provide new insight into the structural properties important for activity of the highly dynamic, trimeric protein disulfide isomerase ScsC from Proteus mirabilis.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
5ID4
 
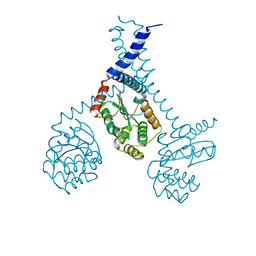 | |
6SCN
 
 | | 33mer structure of the Salmonella flagella MS-ring protein FliF | | 分子名称: | Flagellar M-ring protein | | 著者 | Johnson, S, Fong, Y.H, Deme, J.C, Furlong, E.J, Kuhlen, L, Lea, S.M. | | 登録日 | 2019-07-24 | | 公開日 | 2020-03-18 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Symmetry mismatch in the MS-ring of the bacterial flagellar rotor explains the structural coordination of secretion and rotation.
Nat Microbiol, 5, 2020
|
|
6SD1
 
 | | Structure of the RBM3/collar region of the Salmonella flagella MS-ring protein FliF with 33-fold symmetry applied | | 分子名称: | Flagellar M-ring protein | | 著者 | Johnson, S, Fong, Y.H, Deme, J.C, Furlong, E.J, Kuhlen, L, Lea, S.M. | | 登録日 | 2019-07-26 | | 公開日 | 2020-03-18 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (2.6 Å) | | 主引用文献 | Symmetry mismatch in the MS-ring of the bacterial flagellar rotor explains the structural coordination of secretion and rotation.
Nat Microbiol, 5, 2020
|
|
6SD3
 
 | | 34mer structure of the Salmonella flagella MS-ring protein FliF | | 分子名称: | Flagellar M-ring protein | | 著者 | Johnson, S, Fong, Y.H, Deme, J.C, Furlong, E.J, Kuhlen, L, Lea, S.M. | | 登録日 | 2019-07-26 | | 公開日 | 2020-03-18 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Symmetry mismatch in the MS-ring of the bacterial flagellar rotor explains the structural coordination of secretion and rotation.
Nat Microbiol, 5, 2020
|
|
6SD2
 
 | | Structure of the RBM2inner region of the Salmonella flagella MS-ring protein FliF with 21-fold symmetry applied. | | 分子名称: | Flagellar M-ring protein | | 著者 | Johnson, S, Fong, Y.H, Deme, J.C, Furlong, E.J, Kuhlen, L, Lea, S.M. | | 登録日 | 2019-07-26 | | 公開日 | 2020-03-18 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Symmetry mismatch in the MS-ring of the bacterial flagellar rotor explains the structural coordination of secretion and rotation.
Nat Microbiol, 5, 2020
|
|
6SD4
 
 | | Structure of the RBM3/collar region of the Salmonella flagella MS-ring protein FliF with 34-fold symmetry applied | | 分子名称: | Flagellar M-ring protein | | 著者 | Johnson, S, Fong, Y.H, Deme, J.C, Furlong, E.J, Kuhlen, L, Lea, S.M. | | 登録日 | 2019-07-26 | | 公開日 | 2020-03-18 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Symmetry mismatch in the MS-ring of the bacterial flagellar rotor explains the structural coordination of secretion and rotation.
Nat Microbiol, 5, 2020
|
|
6SD5
 
 | | Structure of the RBM2 inner ring of Salmonella flagella MS-ring protein FliF with 22-fold symmetry applied | | 分子名称: | Flagellar M-ring protein | | 著者 | Johnson, S, Fong, Y.H, Deme, J.C, Furlong, E.J, Kuhlen, L, Lea, S.M. | | 登録日 | 2019-07-26 | | 公開日 | 2020-03-18 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Symmetry mismatch in the MS-ring of the bacterial flagellar rotor explains the structural coordination of secretion and rotation.
Nat Microbiol, 5, 2020
|
|
6TRE
 
 | | Structure of the RBM3/collar region of the Salmonella flagella MS-ring protein FliF with 32-fold symmetry applied | | 分子名称: | Flagellar M-ring protein | | 著者 | Johnson, S, Fong, Y.H, Deme, J.C, Furlong, E.J, Kuhlen, L, Lea, S.M. | | 登録日 | 2019-12-18 | | 公開日 | 2020-03-18 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structure of the bacterial flagellar rotor MS-ring: a minimum inventory/maximum diversity system.
To Be Published
|
|
7NVG
 
 | | Salmonella flagellar basal body refined in C1 map | | 分子名称: | Basal-body rod modification protein FlgD, Flagellar L-ring protein, Flagellar M-ring protein, ... | | 著者 | Johnson, S, Furlong, E, Lea, S.M. | | 登録日 | 2021-03-15 | | 公開日 | 2021-05-05 | | 最終更新日 | 2021-07-14 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Molecular structure of the intact bacterial flagellar basal body.
Nat Microbiol, 6, 2021
|
|
7BK0
 
 | |
7BJ2
 
 | |
7BGL
 
 | | Salmonella LP ring 26 mer refined in C26 map | | 分子名称: | (2~{R},4~{R},5~{R},6~{R})-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-4,5-bis(oxidanyl)oxane-2-carboxylic acid, Flagellar L-ring protein, Flagellar P-ring protein, ... | | 著者 | Johnson, S, Furlong, E, Lea, S.M. | | 登録日 | 2021-01-07 | | 公開日 | 2021-05-05 | | 最終更新日 | 2021-07-14 | | 実験手法 | ELECTRON MICROSCOPY (2.2 Å) | | 主引用文献 | Molecular structure of the intact bacterial flagellar basal body.
Nat Microbiol, 6, 2021
|
|
7BHQ
 
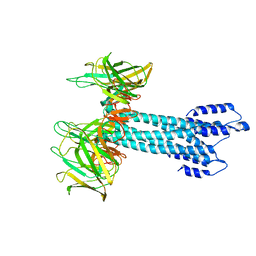 | |
7BIN
 
 | | Salmonella export gate and rod refined in focussed C1 map | | 分子名称: | Flagellar basal body rod protein FlgB, Flagellar basal-body rod protein FlgC, Flagellar basal-body rod protein FlgF, ... | | 著者 | Johnson, S, Furlong, E, Lea, S.M. | | 登録日 | 2021-01-12 | | 公開日 | 2021-05-05 | | 最終更新日 | 2021-06-09 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Molecular structure of the intact bacterial flagellar basal body.
Nat Microbiol, 6, 2021
|
|
7RGV
 
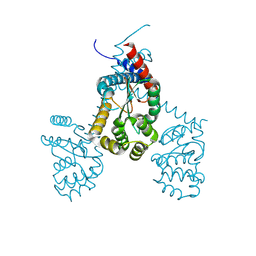 | |
8UPL
 
 | | Cryo-EM structure of a Clockwise locked form of the Salmonella enterica Typhimurium flagellar C-ring, with C34 symmetry applied | | 分子名称: | Flagellar M-ring protein, Flagellar motor switch protein FliG, Flagellar motor switch protein FliM, ... | | 著者 | Johnson, S, Deme, J.C, Lea, S.M. | | 登録日 | 2023-10-22 | | 公開日 | 2024-01-24 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (5.4 Å) | | 主引用文献 | Structural basis of directional switching by the bacterial flagellum.
Nat Microbiol, 9, 2024
|
|
8UOX
 
 | | Cryo-EM structure of a Counterclockwise locked form of the Salmonella enterica Typhimurium flagellar C-ring, with C34 symmetry applied | | 分子名称: | Flagellar M-ring protein, Flagellar motor switch protein FliG, Flagellar motor switch protein FliM, ... | | 著者 | Johnson, S, Deme, J.C, Lea, S.M. | | 登録日 | 2023-10-20 | | 公開日 | 2024-01-24 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (4.6 Å) | | 主引用文献 | Structural basis of directional switching by the bacterial flagellum.
Nat Microbiol, 9, 2024
|
|
8UMD
 
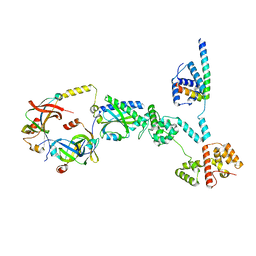 | | Cryo-EM structure of a single subunit of a Counterclockwise-locked form of the Salmonella enterica Typhimurium flagellar C-ring. | | 分子名称: | Flagellar M-ring protein, Flagellar motor switch protein FliG, Flagellar motor switch protein FliM, ... | | 著者 | Johnson, S, Deme, J.C, Lea, S.M. | | 登録日 | 2023-10-17 | | 公開日 | 2024-01-24 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structural basis of directional switching by the bacterial flagellum.
Nat Microbiol, 9, 2024
|
|
8UCS
 
 | |
8UMX
 
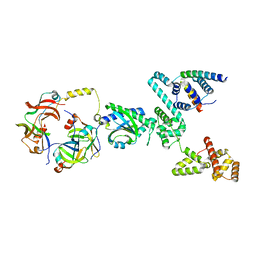 | | Cryo-EM structure of a single subunit of a Clockwise-locked form of the Salmonella enterica Typhimurium flagellar C-ring. | | 分子名称: | Flagellar M-ring protein, Flagellar motor switch protein FliG, Flagellar motor switch protein FliM, ... | | 著者 | Johnson, S, Deme, J.C, Lea, S.M. | | 登録日 | 2023-10-18 | | 公開日 | 2024-01-24 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Structural basis of directional switching by the bacterial flagellum.
Nat Microbiol, 9, 2024
|
|
