8CKP
 
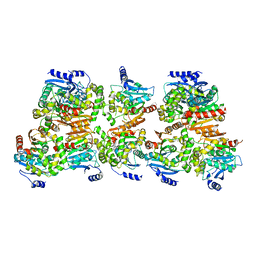 | | X-ray structure of the crystallization-prone form of subfamily III haloalkane dehalogenase DhmeA from Haloferax mediterranei | | 分子名称: | Alpha/beta fold hydrolase, CHLORIDE ION | | 著者 | Marek, M, Chmelova, K, Schenkmayerova, A, Croll, T, Read, R.J, Diederichs, K. | | 登録日 | 2023-02-16 | | 公開日 | 2023-08-30 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (3.31 Å) | | 主引用文献 | Multimeric structure of a subfamily III haloalkane dehalogenase-like enzyme solved by combination of cryo-EM and x-ray crystallography.
Protein Sci., 32, 2023
|
|
8OOH
 
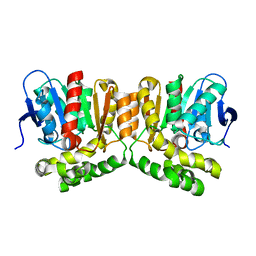 | |
6SP8
 
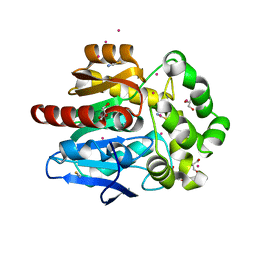 | | Structure of hyperstable haloalkane dehalogenase variant DhaA115 prepared by the 'soak-and-freeze' method under 150 bar of krypton pressure | | 分子名称: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, Haloalkane dehalogenase, ... | | 著者 | Chmelova, K, Markova, K, Damborsky, J, Marek, M. | | 登録日 | 2019-08-31 | | 公開日 | 2020-11-18 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Decoding the intricate network of molecular interactions of a hyperstable engineered biocatalyst.
Chem Sci, 11, 2020
|
|
6SP5
 
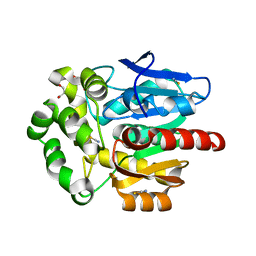 | | Structure of hyperstable haloalkane dehalogenase variant DhaA115 | | 分子名称: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, Haloalkane dehalogenase, ... | | 著者 | Chmelova, K, Markova, K, Damborsky, J, Marek, M. | | 登録日 | 2019-08-30 | | 公開日 | 2020-11-18 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Decoding the intricate network of molecular interactions of a hyperstable engineered biocatalyst.
Chem Sci, 11, 2020
|
|
6XT8
 
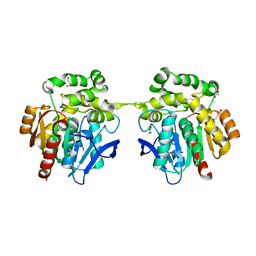 | | Crystal structure of haloalkane dehalogenase variant DhaA115 domain-swapped dimer type-2 | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | 著者 | Markova, K, Damborsky, J, Marek, M. | | 登録日 | 2020-01-15 | | 公開日 | 2021-01-27 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Computational Enzyme Stabilization Can Affect Folding Energy Landscapes and Lead to Catalytically Enhanced Domain-Swapped Dimers
Acs Catalysis, 11, 2021
|
|
6XTC
 
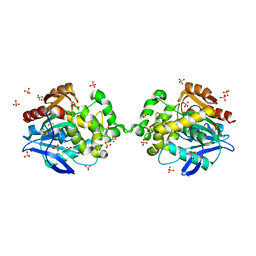 | |
6TY7
 
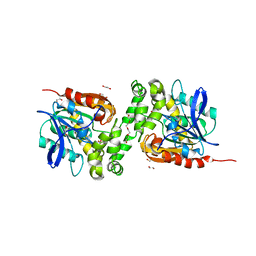 | | Crystal structure of haloalkane dehalogenase variant DhaA115 domain-swapped dimer type-1 | | 分子名称: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, ... | | 著者 | Markova, K, Chaloupkova, R, Damborsky, J, Marek, M. | | 登録日 | 2020-01-15 | | 公開日 | 2021-01-27 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Computational Enzyme Stabilization Can Affect Folding Energy Landscapes and Lead to Catalytically Enhanced Domain-Swapped Dimers
Acs Catalysis, 11, 2021
|
|
