5B0Q
 
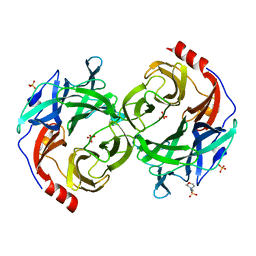 | | beta-1,2-Mannobiose phosphorylase from Listeria innocua - mannose complex | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Lin0857 protein, SULFATE ION, ... | | 著者 | Tsuda, T, Arakawa, T, Fushinobu, S. | | 登録日 | 2015-11-02 | | 公開日 | 2015-12-02 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Characterization and crystal structure determination of beta-1,2-mannobiose phosphorylase from Listeria innocua
Febs Lett., 589, 2015
|
|
5DGQ
 
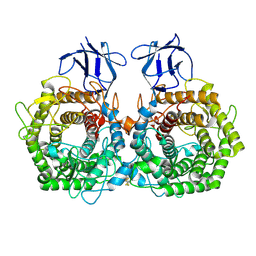 | |
5DGR
 
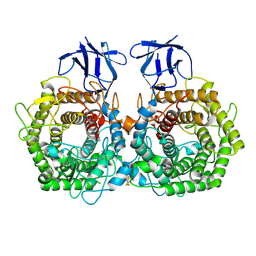 | | Crystal structure of GH9 exo-beta-D-glucosaminidase PBPRA0520, glucosamine complex | | 分子名称: | 2-amino-2-deoxy-beta-D-glucopyranose, Putative endoglucanase-related protein, SODIUM ION | | 著者 | Suzuki, K, Honda, Y, Fushinobu, S. | | 登録日 | 2015-08-28 | | 公開日 | 2015-12-09 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | The crystal structure of an inverting glycoside hydrolase family 9 exo-beta-D-glucosaminidase and the design of glycosynthase.
Biochem.J., 473, 2016
|
|
3WDS
 
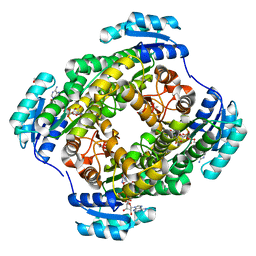 | |
5GX9
 
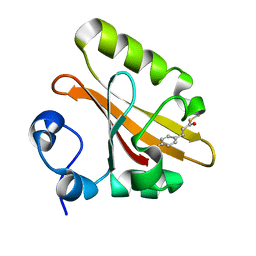 | | PYP mutant - E46Q | | 分子名称: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | 著者 | Yonezawa, K. | | 登録日 | 2016-09-16 | | 公開日 | 2017-08-30 | | 最終更新日 | 2023-11-08 | | 実験手法 | NEUTRON DIFFRACTION (1.493 Å) | | 主引用文献 | Neutron crystallography of photoactive yellow protein reveals unusual protonation state of Arg52 in the crystal
Sci Rep, 7, 2017
|
|
2ZOH
 
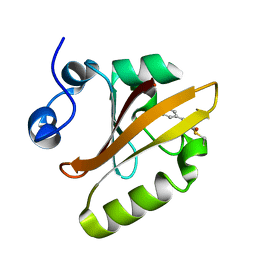 | |
2ZOI
 
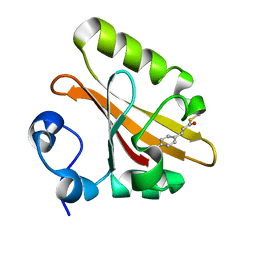 | |
1DEG
 
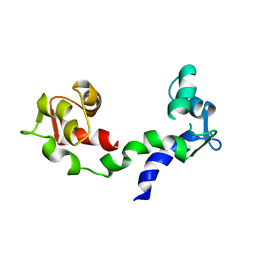 | | THE LINKER OF DES-GLU84 CALMODULIN IS BENT AS SEEN IN THE CRYSTAL STRUCTURE | | 分子名称: | CALCIUM ION, CALMODULIN | | 著者 | Raghunathan, S, Chandross, R, Cheng, B.P, Persechini, A, Sobottk, S.E, Kretsinger, R.H. | | 登録日 | 1993-06-07 | | 公開日 | 1994-05-31 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | The linker of des-Glu84-calmodulin is bent.
Proc.Natl.Acad.Sci.Usa, 90, 1993
|
|
