5H42
 
 | | Crystal Structure of 1,2-beta-oligoglucan phosphorylase from Lachnoclostridium phytofermentans in complex with alpha-d-glucose-1-phosphate | | 分子名称: | 1-O-phosphono-alpha-D-glucopyranose, Uncharacterized protein, alpha-D-glucopyranose | | 著者 | Nakajima, M, Tanaka, N, Furukawa, N, Nihira, T, Kodutsumi, Y, Takahashi, Y, Sugimoto, N, Miyanaga, A, Fushinobu, S, Taguchi, H, Nakai, H. | | 登録日 | 2016-10-28 | | 公開日 | 2017-03-01 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Mechanistic insight into the substrate specificity of 1,2-beta-oligoglucan phosphorylase from Lachnoclostridium phytofermentans
Sci Rep, 7, 2017
|
|
4X5I
 
 | | ecDHFR complexed with folate and NADP+ at 660 MPa | | 分子名称: | BETA-MERCAPTOETHANOL, Dihydrofolate reductase, FOLIC ACID, ... | | 著者 | Yamada, H, Watanabe, N, Nagae, T. | | 登録日 | 2014-12-05 | | 公開日 | 2016-01-13 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | High-pressure protein crystal structure analysis of Escherichia coli dihydrofolate reductase complexed with folate and NADP.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
4WM5
 
 | | High pressure protein crystallography of hen egg white lysozyme at 890 MPa | | 分子名称: | CHLORIDE ION, Lysozyme C, SODIUM ION | | 著者 | Yamada, H, Nagae, T, Watanabe, N. | | 登録日 | 2014-10-08 | | 公開日 | 2015-04-08 | | 最終更新日 | 2020-02-05 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | High-pressure protein crystallography of hen egg-white lysozyme
Acta Crystallogr.,Sect.D, 71, 2015
|
|
1GC0
 
 | | CRYSTAL STRUCTURE OF THE PYRIDOXAL-5'-PHOSPHATE DEPENDENT L-METHIONINE GAMMA-LYASE FROM PSEUDOMONAS PUTIDA | | 分子名称: | METHIONINE GAMMA-LYASE | | 著者 | Motoshima, H, Inagaki, K, Kumasaka, T, Furuichi, M, Inoue, H, Tamura, T, Esaki, N, Soda, K, Tanaka, N, Yamamoto, M, Tanaka, H. | | 登録日 | 2000-07-06 | | 公開日 | 2002-05-08 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystal structure of the pyridoxal 5'-phosphate dependent L-methionine gamma-lyase from Pseudomonas putida.
J.Biochem., 128, 2000
|
|
1A05
 
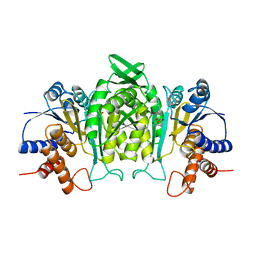 | | CRYSTAL STRUCTURE OF THE COMPLEX OF 3-ISOPROPYLMALATE DEHYDROGENASE FROM THIOBACILLUS FERROOXIDANS WITH 3-ISOPROPYLMALATE | | 分子名称: | 3-ISOPROPYLMALATE DEHYDROGENASE, 3-ISOPROPYLMALIC ACID, MAGNESIUM ION | | 著者 | Imada, K, Inagaki, K, Matsunami, H, Kawaguchi, H, Tanaka, H, Tanaka, N, Namba, K. | | 登録日 | 1997-12-09 | | 公開日 | 1998-06-17 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure of 3-isopropylmalate dehydrogenase in complex with 3-isopropylmalate at 2.0 A resolution: the role of Glu88 in the unique substrate-recognition mechanism.
Structure, 6, 1998
|
|
1WTN
 
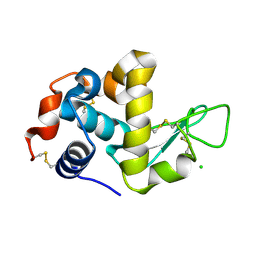 | | The structure of HEW Lysozyme Orthorhombic Crystal Growth under a High Magnetic Field | | 分子名称: | CHLORIDE ION, Lysozyme C | | 著者 | Saijo, S, Yamada, Y, Sato, T, Tanaka, N, Matsui, T, Sazaki, G, Nakajima, K, Matsuura, Y. | | 登録日 | 2004-11-25 | | 公開日 | 2004-12-14 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.13 Å) | | 主引用文献 | Structural consequences of hen egg-white lysozyme orthorhombic crystal growth in a high magnetic field: validation of X-ray diffraction intensity, conformational energy searching and quantitative analysis of B factors and mosaicity.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1UJ0
 
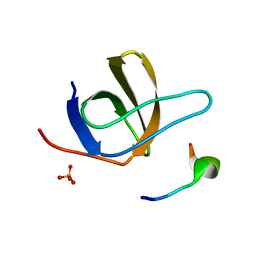 | | Crystal Structure of STAM2 SH3 domain in complex with a UBPY-derived peptide | | 分子名称: | PHOSPHATE ION, deubiquitinating enzyme UBPY, signal transducing adaptor molecule (SH3 domain and ITAM motif) 2 | | 著者 | Kaneko, T, Kumasaka, T, Ganbe, T, Sato, T, Miyazawa, K, Kitamura, N, Tanaka, N. | | 登録日 | 2003-07-24 | | 公開日 | 2003-12-23 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural insight into modest binding of a non-PXXP ligand to the signal transducing adaptor molecule-2 Src homology 3 domain.
J.Biol.Chem., 278, 2003
|
|
1V7Z
 
 | | creatininase-product complex | | 分子名称: | MANGANESE (II) ION, N-[(E)-AMINO(IMINO)METHYL]-N-METHYLGLYCINE, SULFATE ION, ... | | 著者 | Yoshimoto, T, Tanaka, N, Kanada, N, Inoue, T, Nakajima, Y, Haratake, M, Nakamura, K.T, Xu, Y, Ito, K. | | 登録日 | 2003-12-26 | | 公開日 | 2004-01-27 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Crystal structures of creatininase reveal the substrate binding site and provide an insight into the catalytic mechanism
J.Mol.Biol., 337, 2004
|
|
1VBU
 
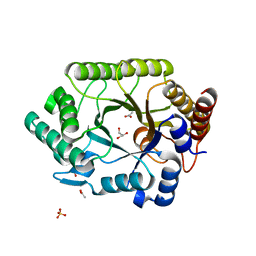 | | Crystal structure of native xylanase 10B from Thermotoga maritima | | 分子名称: | ACETIC ACID, GLYCEROL, SULFATE ION, ... | | 著者 | Ihsanawati, Kumasaka, T, Kaneko, T, Nakamura, S, Tanaka, N. | | 登録日 | 2004-03-02 | | 公開日 | 2005-06-28 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural basis of the substrate subsite and the highly thermal stability of xylanase 10B from Thermotoga maritima MSB8
Proteins, 61, 2005
|
|
1VBR
 
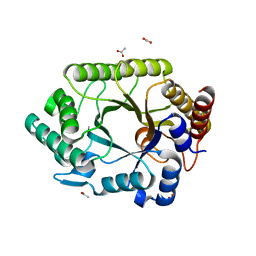 | | Crystal structure of complex xylanase 10B from Thermotoga maritima with xylobiose | | 分子名称: | ACETIC ACID, alpha-D-xylopyranose-(1-4)-beta-D-xylopyranose, endo-1,4-beta-xylanase B | | 著者 | Ihsanawati, Kumasaka, T, Kaneko, T, Nakamura, S, Tanaka, N. | | 登録日 | 2004-03-02 | | 公開日 | 2005-06-28 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural basis of the substrate subsite and the highly thermal stability of xylanase 10B from Thermotoga maritima MSB8
Proteins, 61, 2005
|
|
1WQS
 
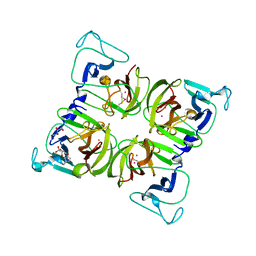 | | Crystal structure of Norovirus 3C-like protease | | 分子名称: | 3C-like protease, D(-)-TARTARIC ACID, L(+)-TARTARIC ACID, ... | | 著者 | Nakamura, K, Someya, Y, Kumasaka, T, Tanaka, N. | | 登録日 | 2004-10-01 | | 公開日 | 2005-10-04 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | A norovirus protease structure provides insights into active and substrate binding site integrity
J.Virol., 79, 2005
|
|
2YZQ
 
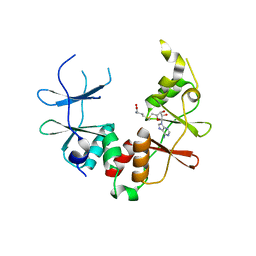 | | Crystal structure of uncharacterized conserved protein from Pyrococcus horikoshii | | 分子名称: | Putative uncharacterized protein PH1780, S-ADENOSYLMETHIONINE | | 著者 | Kanagawa, M, Minami, Y, Watanabe, N, Yokoyama, S, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2007-05-06 | | 公開日 | 2007-11-06 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.63 Å) | | 主引用文献 | Crystal structure of uncharacterized conserved protein from Pyrococcus horikoshii
To be Published
|
|
1WNI
 
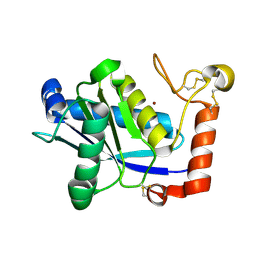 | | Crystal Structure of H2-Proteinase | | 分子名称: | Trimerelysin II, ZINC ION | | 著者 | Kumasaka, T, Yamamoto, M, Moriyama, H, Tanaka, N, Sato, M, Katsube, Y, Yamakawa, Y, Omori-Satoh, T, Iwanaga, S, Ueki, T. | | 登録日 | 2004-08-04 | | 公開日 | 2004-08-17 | | 最終更新日 | 2019-10-09 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structure of H2-proteinase from the venom of Trimeresurus flavoviridis.
J.Biochem., 119, 1996
|
|
1J31
 
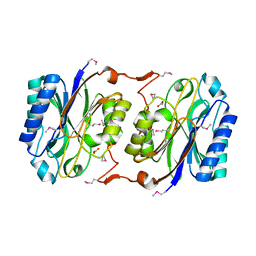 | | Crystal Structure of Hypothetical Protein PH0642 from Pyrococcus horikoshii | | 分子名称: | ACETATE ION, Hypothetical protein PH0642 | | 著者 | Sakai, N, Tajika, Y, Yao, M, Watanabe, N, Tanaka, I. | | 登録日 | 2003-01-16 | | 公開日 | 2004-03-09 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Crystal structure of hypothetical protein PH0642 from Pyrococcus horikoshii at 1.6A resolution.
Proteins, 57, 2004
|
|
1VEC
 
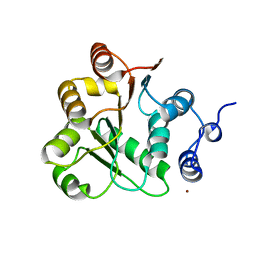 | | Crystal structure of the N-terminal domain of rck/p54, a human DEAD-box protein | | 分子名称: | ATP-dependent RNA helicase p54, L(+)-TARTARIC ACID, ZINC ION | | 著者 | Hogetsu, K, Matsui, T, Yukihiro, Y, Tanaka, M, Sato, T, Kumasaka, T, Tanaka, N. | | 登録日 | 2004-03-29 | | 公開日 | 2004-04-13 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | Structural insight of human DEAD-box protein rck/p54 into its substrate recognition with conformational changes
Genes Cells, 11, 2006
|
|
7CAT
 
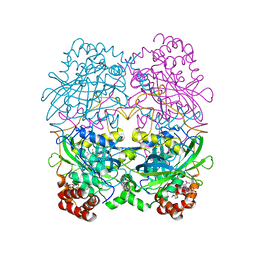 | | The NADPH binding site on beef liver catalase | | 分子名称: | CATALASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Murthy, M.R.N, Reid III, T.J, Sicignano, A, Tanaka, N, Fita, I, Rossmann, M.G. | | 登録日 | 1984-11-15 | | 公開日 | 1985-04-01 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | The NADPH binding site on beef liver catalase.
Proc.Natl.Acad.Sci.USA, 82, 1985
|
|
2JPE
 
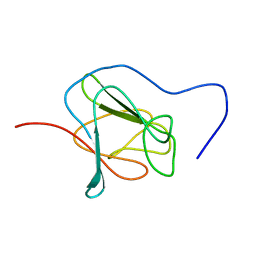 | | FHA domain of NIPP1 | | 分子名称: | Nuclear inhibitor of protein phosphatase 1 | | 著者 | Kumeta, H, Ogura, K, Fujioka, Y, Tanuma, N, Kikuchi, K, Inagaki, F. | | 登録日 | 2007-05-07 | | 公開日 | 2007-05-15 | | 最終更新日 | 2024-05-08 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The NMR structure of the NIPP1 FHA domain.
J.Biomol.Nmr, 40, 2008
|
|
1IPD
 
 | | THREE-DIMENSIONAL STRUCTURE OF A HIGHLY THERMOSTABLE ENZYME, 3-ISOPROPYLMALATE DEHYDROGENASE OF THERMUS THERMOPHILUS AT 2.2 ANGSTROMS RESOLUTION | | 分子名称: | 3-ISOPROPYLMALATE DEHYDROGENASE, SULFATE ION | | 著者 | Imada, K, Sato, M, Tanaka, N, Katsube, Y, Matsuura, Y, Oshima, T. | | 登録日 | 1992-01-29 | | 公開日 | 1993-10-31 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Three-dimensional structure of a highly thermostable enzyme, 3-isopropylmalate dehydrogenase of Thermus thermophilus at 2.2 A resolution.
J.Mol.Biol., 222, 1991
|
|
2RQO
 
 | | Solution structure of Polytheonamide B | | 分子名称: | polytheonamide B | | 著者 | Hamada, N, Matsunaga, S, Fujiwara, M, Fujjita, K, Hirota, H, Schmucki, R, Guntert, P, Fusetani, N. | | 登録日 | 2009-09-03 | | 公開日 | 2010-09-15 | | 最終更新日 | 2023-11-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution Structure of Polytheonamide B, a Highly Cytotoxic Nonribosomal Polypeptide from Marine Sponge
J.Am.Chem.Soc., 2010
|
|
1OSJ
 
 | | STRUCTURE OF 3-ISOPROPYLMALATE DEHYDROGENASE | | 分子名称: | 3-ISOPROPYLMALATE DEHYDROGENASE | | 著者 | Qu, C, Akanuma, S, Moriyama, H, Tanaka, N, Oshima, T. | | 登録日 | 1996-10-22 | | 公開日 | 1997-01-27 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | A mutation at the interface between domains causes rearrangement of domains in 3-isopropylmalate dehydrogenase.
Protein Eng., 10, 1997
|
|
1UIR
 
 | | Crystal Structure of Polyamine Aminopropyltransfease from Thermus thermophilus | | 分子名称: | Polyamine Aminopropyltransferase | | 著者 | Ganbe, T, Ohnuma, M, Sato, T, Kumasaka, T, Oshima, T, Tanaka, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2003-07-18 | | 公開日 | 2003-08-05 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structures and enzymatic properties of a triamine/agmatine aminopropyltransferase from Thermus thermophilus
J.Mol.Biol., 408, 2011
|
|
7DKD
 
 | | Stenotrophomonas maltophilia DPP7 in complex with Asn-Tyr | | 分子名称: | ASPARAGINE, Dipeptidyl-peptidase, GLYCEROL, ... | | 著者 | Sakamoto, Y, Nakamura, A, Suzuki, Y, Honma, N, Roppongi, S, Kushibiki, C, Yonezawa, N, Takahashi, M, Shida, Y, Gouda, H, Nonaka, T, Ogasawara, W, Tanaka, N. | | 登録日 | 2020-11-23 | | 公開日 | 2021-11-03 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | Structural basis for an exceptionally strong preference for asparagine residue at the S2 subsite of Stenotrophomonas maltophilia dipeptidyl peptidase 7.
Sci Rep, 11, 2021
|
|
7DKC
 
 | | Stenotrophomonas maltophilia DPP7 in complex with Tyr-Tyr | | 分子名称: | Dipeptidyl-peptidase, GLYCEROL, TYROSINE | | 著者 | Sakamoto, Y, Nakamura, A, Suzuki, Y, Honma, N, Roppongi, S, Kushibiki, C, Yonezawa, N, Takahashi, M, Shida, Y, Gouda, H, Nonaka, T, Ogasawara, W, Tanaka, N. | | 登録日 | 2020-11-23 | | 公開日 | 2021-11-03 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.86 Å) | | 主引用文献 | Structural basis for an exceptionally strong preference for asparagine residue at the S2 subsite of Stenotrophomonas maltophilia dipeptidyl peptidase 7.
Sci Rep, 11, 2021
|
|
7DKE
 
 | | Stenotrophomonas maltophilia DPP7 in complex with Phe-Tyr | | 分子名称: | Dipeptidyl-peptidase, GLYCEROL, PHENYLALANINE, ... | | 著者 | Sakamoto, Y, Nakamura, A, Suzuki, Y, Honma, N, Roppongi, S, Kushibiki, C, Yonezawa, N, Takahashi, M, Shida, Y, Gouda, H, Nonaka, T, Ogasawara, W, Tanaka, N. | | 登録日 | 2020-11-23 | | 公開日 | 2021-11-03 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.91 Å) | | 主引用文献 | Structural basis for an exceptionally strong preference for asparagine residue at the S2 subsite of Stenotrophomonas maltophilia dipeptidyl peptidase 7.
Sci Rep, 11, 2021
|
|
7DKB
 
 | | Stenotrophomonas maltophilia DPP7 in complex with Val-Tyr | | 分子名称: | Dipeptidyl-peptidase, TYROSINE, VALINE | | 著者 | Sakamoto, Y, Nakamura, A, Suzuki, Y, Honma, N, Roppongi, S, Kushibiki, C, Yonezawa, N, Takahashi, M, Shida, Y, Gouda, H, Nonaka, T, Ogasawara, W, Tanaka, N. | | 登録日 | 2020-11-23 | | 公開日 | 2021-11-03 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | Structural basis for an exceptionally strong preference for asparagine residue at the S2 subsite of Stenotrophomonas maltophilia dipeptidyl peptidase 7.
Sci Rep, 11, 2021
|
|
