7CRT
 
 | | Structure of Chloride ion pumping rhodopsin (ClR) with NTQ motif 100 ps after light activation (0.17mJ/mm2) | | 分子名称: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | 著者 | Yun, J.H, Liu, H, Lee, W.T, Schmidt, M. | | 登録日 | 2020-08-14 | | 公開日 | 2021-04-14 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Early-stage dynamics of chloride ion-pumping rhodopsin revealed by a femtosecond X-ray laser.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7CRS
 
 | | Structure of Chloride ion pumping rhodopsin (ClR) with NTQ motif 100 ps after light activation (0.90mJ/mm2) | | 分子名称: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | 著者 | Yun, J.H, Liu, H, Lee, W.T, Schmidt, M. | | 登録日 | 2020-08-14 | | 公開日 | 2021-04-14 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Early-stage dynamics of chloride ion-pumping rhodopsin revealed by a femtosecond X-ray laser.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7CUE
 
 | |
7CUD
 
 | |
1JEU
 
 | |
1JEV
 
 | |
1JET
 
 | |
7CRJ
 
 | | Dark State Structure of Chloride ion pumping rhodopsin (ClR) with NTQ motif | | 分子名称: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | 著者 | Yun, J.H, Liu, H, Lee, W.T, Schmidt, M. | | 登録日 | 2020-08-13 | | 公開日 | 2020-09-16 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Early-stage dynamics of chloride ion-pumping rhodopsin revealed by a femtosecond X-ray laser.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
1UEB
 
 | | Crystal structure of translation elongation factor P from Thermus thermophilus HB8 | | 分子名称: | elongation factor P | | 著者 | Hanawa-Suetsugu, K, Sekine, S, Sakai, H, Hori-Takemoto, C, Terada, T, Kuramitsu, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2003-05-09 | | 公開日 | 2004-05-25 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Crystal structure of elongation factor P from Thermus thermophilus HB8
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1UEK
 
 | | Crystal structure of 4-(cytidine 5'-diphospho)-2C-methyl-D-erythritol kinase | | 分子名称: | 4-(cytidine 5'-diphospho)-2C-methyl-D-erythritol kinase | | 著者 | Wada, T, Kuramitsu, S, Yokoyama, S, Tame, J.R.H, Park, S.Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2003-05-17 | | 公開日 | 2003-06-17 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystal Structure of 4-(Cytidine 5'-diphospho)-2-C-methyl-D-erythritol kinase, an Enzyme in the Non-mevalonate Pathway of Isoprenoid Synthesis.
J.Biol.Chem., 278, 2003
|
|
6I38
 
 | |
6I3A
 
 | | Crystal structure of v22Pizza6-AYW, a circularly permuted designer protein | | 分子名称: | BROMIDE ION, v22Pizza6-AYW | | 著者 | Mylemans, B, Noguchi, H, Deridder, E, Voet, A.R.D. | | 登録日 | 2018-11-05 | | 公開日 | 2019-11-20 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Influence of circular permutations on the structure and stability of a six-fold circular symmetric designer protein.
Protein Sci., 2020
|
|
6JYA
 
 | | Structure of dark-state marine bacterial chloride importer, NM-R3, with CW laser (ND-10%) at 95K. | | 分子名称: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | 著者 | Yun, J.H, Ohki, M, Park, S.Y, Lee, W. | | 登録日 | 2019-04-26 | | 公開日 | 2020-03-04 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.803 Å) | | 主引用文献 | Pumping mechanism of NM-R3, a light-driven bacterial chloride importer in the rhodopsin family.
Sci Adv, 6, 2020
|
|
6JYF
 
 | | Structure of light-state marine bacterial chloride importer, NM-R3, with Pulse laser (ND-1%) at 140K. | | 分子名称: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | 著者 | Yun, J.H, Ohki, M, Park, S.Y, Lee, W. | | 登録日 | 2019-04-26 | | 公開日 | 2020-03-04 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.004 Å) | | 主引用文献 | Pumping mechanism of NM-R3, a light-driven bacterial chloride importer in the rhodopsin family.
Sci Adv, 6, 2020
|
|
6JYE
 
 | | Structure of dark-state marine bacterial chloride importer, NM-R3, with Pulse laser (ND-1%) at 140K. | | 分子名称: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | 著者 | Yun, J.H, Ohki, M, Park, S.Y, Lee, W. | | 登録日 | 2019-04-26 | | 公開日 | 2020-03-04 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Pumping mechanism of NM-R3, a light-driven bacterial chloride importer in the rhodopsin family.
Sci Adv, 6, 2020
|
|
6I37
 
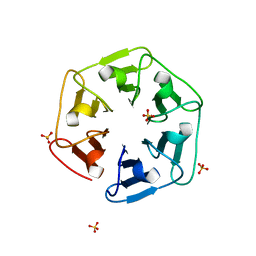 | | Crystal structure of nv1Pizza6-AYW, a circularly permuted designer protein | | 分子名称: | SULFATE ION, nv1Pizza6-AYW | | 著者 | Mylemans, B, Noguchi, H, Deridder, E, Voet, A.R.D. | | 登録日 | 2018-11-05 | | 公開日 | 2019-11-20 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.54 Å) | | 主引用文献 | Influence of circular permutations on the structure and stability of a six-fold circular symmetric designer protein.
Protein Sci., 2020
|
|
6I39
 
 | | Crystal structure of v31Pizza6-AYW, a circularly permuted designer protein | | 分子名称: | MAGNESIUM ION, v31Pizza6-AYW | | 著者 | Mylemans, B, Noguchi, H, Deridder, E, Voet, A.R.D. | | 登録日 | 2018-11-05 | | 公開日 | 2019-11-20 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.05 Å) | | 主引用文献 | Influence of circular permutations on the structure and stability of a six-fold circular symmetric designer protein.
Protein Sci., 2020
|
|
6JY6
 
 | | Structure of dark-state marine bacterial chloride importer, NM-R3, with Pulse laser (ND-1%) at 95K. | | 分子名称: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | 著者 | Yun, J.H, Ohki, M, Park, S.Y, Lee, W. | | 登録日 | 2019-04-26 | | 公開日 | 2020-03-04 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Pumping mechanism of NM-R3, a light-driven bacterial chloride importer in the rhodopsin family.
Sci Adv, 6, 2020
|
|
6JYC
 
 | | Structure of dark-state marine bacterial chloride importer, NM-R3, with CW laser (ND-30%) at 95K. | | 分子名称: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | 著者 | Yun, J.H, Ohki, M, Park, S.Y, Lee, W. | | 登録日 | 2019-04-26 | | 公開日 | 2020-03-04 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.892 Å) | | 主引用文献 | Pumping mechanism of NM-R3, a light-driven bacterial chloride importer in the rhodopsin family.
Sci Adv, 6, 2020
|
|
6JY8
 
 | | Structure of dark-state marine bacterial chloride importer, NM-R3, with CW laser (ND-3%) at 95K. | | 分子名称: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | 著者 | Yun, J.H, Ohki, M, Park, S.Y, Lee, W. | | 登録日 | 2019-04-26 | | 公開日 | 2020-03-04 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Pumping mechanism of NM-R3, a light-driven bacterial chloride importer in the rhodopsin family.
Sci Adv, 6, 2020
|
|
6JY7
 
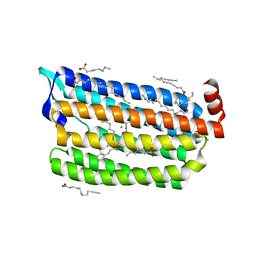 | | Structure of light-state marine bacterial chloride importer, NM-R3, with Pulse laser (ND-1%) at 95K. | | 分子名称: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | 著者 | Yun, J.H, Ohki, M, Park, S.Y, Lee, W. | | 登録日 | 2019-04-26 | | 公開日 | 2020-03-04 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Pumping mechanism of NM-R3, a light-driven bacterial chloride importer in the rhodopsin family.
Sci Adv, 6, 2020
|
|
6JYD
 
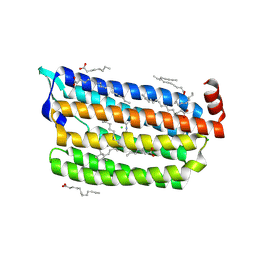 | | Structure of light-state marine bacterial chloride importer, NM-R3, with CW laser (ND-30%) at 95K. | | 分子名称: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | 著者 | Yun, J.H, Ohki, M, Park, S.Y, Lee, W. | | 登録日 | 2019-04-26 | | 公開日 | 2020-03-04 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.007 Å) | | 主引用文献 | Pumping mechanism of NM-R3, a light-driven bacterial chloride importer in the rhodopsin family.
Sci Adv, 6, 2020
|
|
6JY9
 
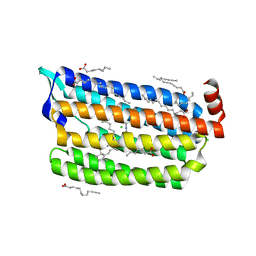 | | Structure of light-state marine bacterial chloride importer, NM-R3, with CW laser (ND-3%) at 95K. | | 分子名称: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | 著者 | Yun, J.H, Ohki, M, Park, S.Y, Lee, W. | | 登録日 | 2019-04-26 | | 公開日 | 2020-03-04 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Pumping mechanism of NM-R3, a light-driven bacterial chloride importer in the rhodopsin family.
Sci Adv, 6, 2020
|
|
6JYB
 
 | | Structure of light-state marine bacterial chloride importer, NM-R3, with CW laser (ND-10%) at 95K. | | 分子名称: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | 著者 | Yun, J.H, Ohki, M, Park, S.Y, Lee, W. | | 登録日 | 2019-04-26 | | 公開日 | 2020-03-04 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.802 Å) | | 主引用文献 | Pumping mechanism of NM-R3, a light-driven bacterial chloride importer in the rhodopsin family.
Sci Adv, 6, 2020
|
|
6I3B
 
 | |
