2JAS
 
 | | Structure of deoxyadenosine kinase from M.mycoides with bound dATP | | 分子名称: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, DEOXYGUANOSINE KINASE, MAGNESIUM ION | | 著者 | Welin, M, Wang, L, Eriksson, S, Eklund, H. | | 登録日 | 2006-11-30 | | 公開日 | 2007-01-03 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structure-Function Analysis of a Bacterial Deoxyadenosine Kinase Reveals the Basis for Substrate Specificity.
J.Mol.Biol., 366, 2007
|
|
1BE1
 
 | | GLUTAMATE MUTASE (B12-BINDING SUBUNIT), NMR, MINIMIZED AVERAGE STRUCTURE | | 分子名称: | GLUTAMATE MUTASE | | 著者 | Tollinger, M, Konrat, R, Hilbert, B.H, Marsh, E.N.G, Kraeutler, B. | | 登録日 | 1998-05-19 | | 公開日 | 1998-08-26 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | How a protein prepares for B12 binding: structure and dynamics of the B12-binding subunit of glutamate mutase from Clostridium tetanomorphum
Structure, 6, 1998
|
|
2JFS
 
 | | Crystal structure of the PPM Ser-Thr phosphatase MsPP from Mycobacterium smegmatis in complex with cacodylate | | 分子名称: | CACODYLATE ION, GLYCEROL, MANGANESE (II) ION, ... | | 著者 | Bellinzoni, M, Wehenkel, A, Shepard, W, Alzari, P.M. | | 登録日 | 2007-02-04 | | 公開日 | 2007-07-24 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Insights Into the Mechanism of Ppm Ser/Thr Phosphatases from the Atomic Resolution Structures of a Mycobacterial Enzyme
Structure, 15, 2007
|
|
2UZ3
 
 | | Crystal Structure of Thymidine Kinase with dTTP from U. urealyticum | | 分子名称: | MAGNESIUM ION, THYMIDINE KINASE, THYMIDINE-5'-TRIPHOSPHATE, ... | | 著者 | Welin, M, Kosinska, U, Mikkelsen, N.E, Carnrot, C, Zhu, C, Wang, L, Eriksson, S, Munch-Petersen, B, Eklund, H. | | 登録日 | 2007-04-24 | | 公開日 | 2007-05-29 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structures of Thymidine Kinase 1 of Human and Mycoplasmic Origin
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
2JFT
 
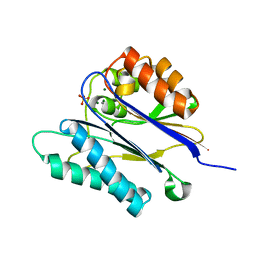 | | Crystal structure of the PPM Ser-Thr phosphatase MsPP from Mycobacterium smegmatis in complex with sulfate | | 分子名称: | MAGNESIUM ION, MANGANESE (II) ION, SER-THR PHOSPHATASE MSPP, ... | | 著者 | Bellinzoni, M, Wehenkel, A, Shepard, W, Alzari, P.M. | | 登録日 | 2007-02-04 | | 公開日 | 2007-07-24 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.08 Å) | | 主引用文献 | Insights Into the Mechanism of Ppm Ser/Thr Phosphatases from the Atomic Resolution Structures of a Mycobacterial Enzyme
Structure, 15, 2007
|
|
2JFR
 
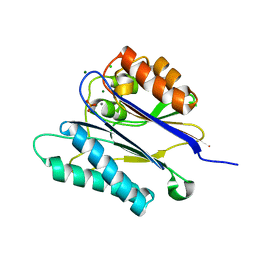 | | Crystal structure of the PPM Ser-Thr phosphatase MsPP from Mycobacterium smegmatis in complex with phosphate at 0.83 A resolution | | 分子名称: | CHLORIDE ION, MAGNESIUM ION, MANGANESE (II) ION, ... | | 著者 | Bellinzoni, M, Wehenkel, A, Shepard, W, Alzari, P.M. | | 登録日 | 2007-02-04 | | 公開日 | 2007-07-24 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (0.83 Å) | | 主引用文献 | Insights Into the Mechanism of Ppm Ser/Thr Phosphatases from the Atomic Resolution Structures of a Mycobacterial Enzyme
Structure, 15, 2007
|
|
2V57
 
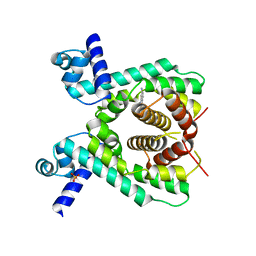 | | Crystal structure of the TetR-like transcriptional regulator LfrR from Mycobacterium smegmatis in complex with proflavine | | 分子名称: | ISOPROPYL ALCOHOL, PROFLAVIN, SULFATE ION, ... | | 著者 | Bellinzoni, M, Buroni, S, Riccardi, G, De Rossi, E, Alzari, P.M. | | 登録日 | 2008-10-02 | | 公開日 | 2009-10-20 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural Plasticity and Distinct Drug-Binding Modes of Lfrr, a Mycobacterial Efflux Pump Regulator.
J.Bacteriol., 191, 2009
|
|
2WGB
 
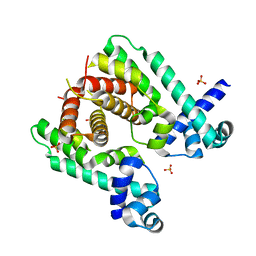 | | Crystal structure of the TetR-like transcriptional regulator LfrR from Mycobacterium smegmatis | | 分子名称: | SULFATE ION, TETR FAMILY TRANSCRIPTIONAL REPRESSOR LFRR | | 著者 | Bellinzoni, M, Buroni, S, Riccardi, G, De Rossi, E, Alzari, P.M. | | 登録日 | 2009-04-16 | | 公開日 | 2009-10-20 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural Plasticity and Distinct Drug-Binding Modes of Lfrr, a Mycobacterial Efflux Pump Regulator.
J.Bacteriol., 191, 2009
|
|
1MKS
 
 | |
8H6I
 
 | | The crystal structure of SARS-CoV-2 3C-like protease Double Mutant (L50F and E166V) in complex with a traditional Chinese Medicine Inhibitors | | 分子名称: | (1beta,6beta,7beta,8alpha,9beta,10alpha,13alpha,14R,16beta)-1,6,7,14-tetrahydroxy-7,20-epoxykauran-15-one, 3C-like proteinase nsp5 | | 著者 | Lin, M, Liu, X. | | 登録日 | 2022-10-17 | | 公開日 | 2023-10-11 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
8H7K
 
 | |
8H5F
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) L167F Mutant in Complex with Inhibitor Nirmatrelvir | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | 著者 | Lin, M, Liu, X. | | 登録日 | 2022-10-13 | | 公開日 | 2023-10-11 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
8H51
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) Double Mutant (T21I and E166V) in Complex with Inhibitor Nirmatrelvir | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | 著者 | Lin, M, Liu, X. | | 登録日 | 2022-10-11 | | 公開日 | 2023-10-11 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.18 Å) | | 主引用文献 | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
8H57
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) A193P Mutant in Complex with Inhibitor Nirmatrelvir | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | 著者 | Lin, M, Liu, X. | | 登録日 | 2022-10-12 | | 公開日 | 2023-10-11 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
8H5P
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) Double Mutant (L50F and E166V) in Complex with Inhibitor Nirmatrelvir | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | 著者 | Lin, M, Liu, X. | | 登録日 | 2022-10-13 | | 公開日 | 2023-10-11 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.67 Å) | | 主引用文献 | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
8H6N
 
 | | Crystal structure of SARS-CoV-2 main protease (Mpro) Mutant (T21I) in complex with protease inhibitor Nirmatrelvir | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 2-(diethylamino)-N-(2,6-dimethylphenyl)ethanamide, 3C-like proteinase nsp5 | | 著者 | Lin, M, Liu, X. | | 登録日 | 2022-10-18 | | 公開日 | 2023-10-11 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
8H7W
 
 | | Crystal structure of SARS-CoV-2 main protease (Mpro) Mutant (S144A) in complex with protease inhibitor Nirmatrelvir | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | 著者 | Lin, M, Liu, X. | | 登録日 | 2022-10-21 | | 公開日 | 2023-10-11 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
8H82
 
 | | Crystal structure of SARS-CoV-2 main protease (Mpro) Mutant (E166V) in complex with protease inhibitor Nirmatrelvir | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | 著者 | Lin, M, Liu, X. | | 登録日 | 2022-10-21 | | 公開日 | 2023-10-11 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
8HBK
 
 | | The crystal structure of SARS-CoV-2 3CL protease in complex with Ensitrelvir | | 分子名称: | 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | | 著者 | Lin, M. | | 登録日 | 2022-10-29 | | 公開日 | 2023-10-11 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
8H4Y
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) F140L Mutant in Complex with Inhibitor Nirmatrelvir | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | 著者 | Lin, M, Liu, X. | | 登録日 | 2022-10-11 | | 公開日 | 2023-10-11 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
6QRO
 
 | | Crystal structure of Tannerella forsythia glutaminyl cyclase | | 分子名称: | Glutamine cyclotransferase, SULFATE ION, ZINC ION | | 著者 | Linnert, M, Piechotta, A, Parthier, C, Taudte, N, Kolenko, P, Rahfeld, J, Potempa, J, Stubbs, M.T. | | 登録日 | 2019-02-19 | | 公開日 | 2019-03-06 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Mammalian-like type II glutaminyl cyclases in Porphyromonas gingivalis and other oral pathogenic bacteria as targets for treatment of periodontitis.
J.Biol.Chem., 296, 2021
|
|
6QQL
 
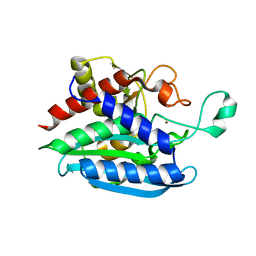 | | Crystal structure of Porphyromonas gingivalis glutaminyl cyclase | | 分子名称: | Glutamine cyclotransferase, ZINC ION | | 著者 | Linnert, M, Piechotta, A, Parthier, C, Taudte, N, Kolenko, P, Rahfeld, J, Potempa, J, Stubbs, M.T. | | 登録日 | 2019-02-18 | | 公開日 | 2019-03-06 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.814 Å) | | 主引用文献 | Mammalian-like type II glutaminyl cyclases in Porphyromonas gingivalis and other oral pathogenic bacteria as targets for treatment of periodontitis.
J.Biol.Chem., 296, 2021
|
|
4TNC
 
 | |
5K59
 
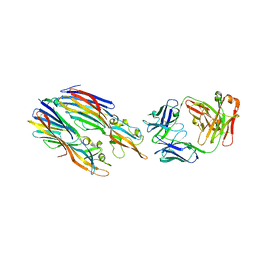 | | Crystal structure of LukGH from Staphylococcus aureus in complex with a neutralising antibody | | 分子名称: | CHLORIDE ION, Fab heavy chain, Fab light chain, ... | | 著者 | Welin, M, Logan, D.T, Badarau, A, Mirkina, I, Zauner, G, Dolezilkova, I, Nagy, E. | | 登録日 | 2016-05-23 | | 公開日 | 2016-08-10 | | 最終更新日 | 2018-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.84 Å) | | 主引用文献 | Context matters: The importance of dimerization-induced conformation of the LukGH leukocidin of Staphylococcus aureus for the generation of neutralizing antibodies.
Mabs, 8, 2016
|
|
6NSV
 
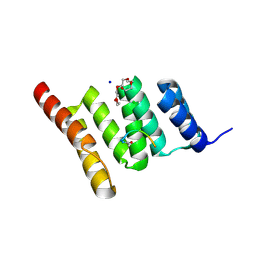 | | Crystal structure of the human CHIP TPR domain in complex with a 5mer acetylated optimized peptide | | 分子名称: | ACE-LEU-TRP-TRP-PRO-ASP, CHLORIDE ION, E3 ubiquitin-protein ligase CHIP, ... | | 著者 | Basu, K, Ravalin, M, Bohn, M.-F, Craik, C.S, Gestwicki, J.E. | | 登録日 | 2019-01-25 | | 公開日 | 2019-07-31 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.305 Å) | | 主引用文献 | Specificity for latent C termini links the E3 ubiquitin ligase CHIP to caspases.
Nat.Chem.Biol., 15, 2019
|
|
