4JS9
 
 | | Structural Characterization of Inducible Nitric Oxide Synthase Substituted With Mesoheme | | 分子名称: | 5,6,7,8-TETRAHYDROBIOPTERIN, Mesoheme, Nitric oxide synthase, ... | | 著者 | Hannibal, L, Page, R.C, Bolisetty, K, Yu, Z, Misra, S, Stuehr, D.J. | | 登録日 | 2013-03-22 | | 公開日 | 2014-04-23 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.784 Å) | | 主引用文献 | Kinetic and Structural Characterization of Inducible Nitric Oxide Synthase Substituted With Mesoheme
To be Published
|
|
1HQ3
 
 | | CRYSTAL STRUCTURE OF THE HISTONE-CORE-OCTAMER IN KCL/PHOSPHATE | | 分子名称: | CHLORIDE ION, HISTONE H2A-IV, HISTONE H2B, ... | | 著者 | Chantalat, L, Nicholson, J.M, Lambert, S.J, Reid, A.J, Donovan, M.J, Reynolds, C.D, Wood, C.M, Baldwin, J.P. | | 登録日 | 2000-12-14 | | 公開日 | 2001-01-24 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Structure of the histone-core octamer in KCl/phosphate crystals at 2.15 A resolution.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1E31
 
 | | SURVIVIN DIMER H. SAPIENS | | 分子名称: | APOPTOSIS INHIBITOR SURVIVIN, COBALT (II) ION, ZINC ION | | 著者 | Chantalat, L, Skoufias, D.A, Margolis, R.L, Dideberg, O. | | 登録日 | 2000-06-04 | | 公開日 | 2001-01-03 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.71 Å) | | 主引用文献 | Crystal Structure of Human Survivin Reveals a Bow Tie-Shaped Dimer with Two Unusual Alpha-Helical Extensions
Mol.Cell, 6, 2000
|
|
1DXK
 
 | |
7XMK
 
 | | Crystal structure of human RIPK1 kinase domain in complex with compound SKLB923 | | 分子名称: | 5-[2-(cyclopropylcarbonylamino)-[1,2,4]triazolo[1,5-a]pyridin-7-yl]-N-[(1S)-1-(3-fluorophenyl)ethyl]-1-methyl-indole-3-carboxamide, IODIDE ION, Receptor-interacting serine/threonine-protein kinase 1 | | 著者 | Zhang, L, Wang, Y, Li, Y, Yang, S. | | 登録日 | 2022-04-26 | | 公開日 | 2023-04-26 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (2.376 Å) | | 主引用文献 | From Hit to Lead: Structure-Based Optimization of Novel Selective Inhibitors of Receptor-Interacting Protein Kinase 1 (RIPK1) for the Treatment of Inflammatory Diseases.
J.Med.Chem., 67, 2024
|
|
2PQE
 
 | | Solution structure of proline-free mutant of staphylococcal nuclease | | 分子名称: | Thermonuclease | | 著者 | Shan, L, Tong, Y, Xie, T, Wang, M, Wang, J. | | 登録日 | 2007-05-01 | | 公開日 | 2007-06-19 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Restricted backbone conformational and motional flexibilities of loops containing peptidyl-proline bonds dominate the enzyme activity of staphylococcal nuclease.
Biochemistry, 46, 2007
|
|
1HGU
 
 | | HUMAN GROWTH HORMONE | | 分子名称: | HUMAN GROWTH HORMONE | | 著者 | Chantalat, L, Jones, N, Korber, F, Navaza, J, Pavlovsky, A.G. | | 登録日 | 1995-05-11 | | 公開日 | 1995-12-07 | | 最終更新日 | 2019-08-14 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | THE CRYSTAL-STRUCTURE OF WILD-TYPE GROWTH-HORMONE AT 2.5 ANGSTROM RESOLUTION.
Protein Pept.Lett., 2, 1995
|
|
4NXL
 
 | | Dibenzothiophene monooxygenase (DszC) from Rhodococcus erythropolis | | 分子名称: | DszC | | 著者 | Zhang, L, Duan, X, Li, X, Rao, Z. | | 登録日 | 2013-12-09 | | 公開日 | 2014-07-23 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural insights into the stabilization of active, tetrameric DszC by its C-terminus.
Proteins, 82, 2014
|
|
7VTY
 
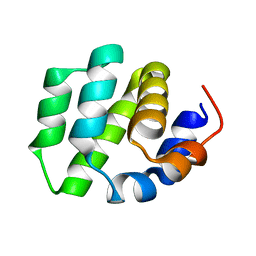 | | de novo designed protein | | 分子名称: | de novo designed protein | | 著者 | Zhang, L. | | 登録日 | 2021-10-31 | | 公開日 | 2022-06-08 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Rotamer-free protein sequence design based on deep learning and self-consistency.
Nat Comput Sci, 2023
|
|
7VQV
 
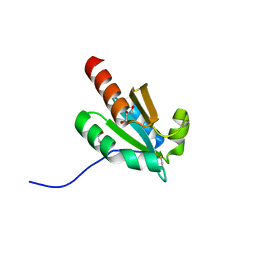 | | de novo design based on 1r26 | | 分子名称: | GLYCEROL, de novo designed protein | | 著者 | Zhang, L. | | 登録日 | 2021-10-20 | | 公開日 | 2022-06-08 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.53 Å) | | 主引用文献 | Rotamer-free protein sequence design based on deep learning and self-consistency.
Nat Comput Sci, 2023
|
|
7VQL
 
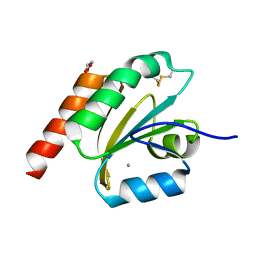 | | de novo designed based on 1r26 | | 分子名称: | AMMONIUM ION, GLYCEROL, de novo designed protein | | 著者 | Zhang, L. | | 登録日 | 2021-10-20 | | 公開日 | 2022-06-08 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Rotamer-free protein sequence design based on deep learning and self-consistency.
Nat Comput Sci, 2023
|
|
7VU4
 
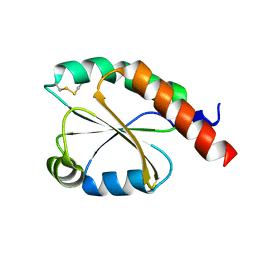 | | de novo design based on 1r26 | | 分子名称: | de novo design protein | | 著者 | Zhang, L. | | 登録日 | 2021-11-01 | | 公開日 | 2022-06-08 | | 最終更新日 | 2023-06-28 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Rotamer-free protein sequence design based on deep learning and self-consistency.
Nat Comput Sci, 2023
|
|
7VQW
 
 | | de novo designed protein based on 1r26 | | 分子名称: | de novo designed protein | | 著者 | Zhang, L. | | 登録日 | 2021-10-20 | | 公開日 | 2022-06-08 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Rotamer-free protein sequence design based on deep learning and self-consistency.
Nat Comput Sci, 2023
|
|
5VFA
 
 | | RitR Mutant - C128D | | 分子名称: | Response regulator | | 著者 | Han, L, Silvaggi, N.R. | | 登録日 | 2017-04-07 | | 公開日 | 2018-04-11 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.452 Å) | | 主引用文献 | RitR is an archetype for a novel family of redox sensors in the streptococci that has evolved from two-component response regulators and is required for pneumococcal colonization.
PLoS Pathog., 14, 2018
|
|
7SLA
 
 | | CryoEM structure of SGLT1 at 3.15 Angstrom resolution | | 分子名称: | CHOLESTEROL HEMISUCCINATE, Sodium/glucose cotransporter 1, nanobody Nb1 | | 著者 | Qu, Q, Han, L, Panova, O, Feng, L, Skiniotis, G. | | 登録日 | 2021-10-23 | | 公開日 | 2021-12-15 | | 最終更新日 | 2022-01-26 | | 実験手法 | ELECTRON MICROSCOPY (3.15 Å) | | 主引用文献 | Structure and mechanism of the SGLT family of glucose transporters.
Nature, 601, 2022
|
|
7SL9
 
 | | CryoEM structure of SMCT1 | | 分子名称: | Sodium-coupled monocarboxylate transporter 1, butanoic acid, nanobody Nb2 | | 著者 | Qu, Q, Han, L, Panova, O, Feng, L, Skiniotis, G. | | 登録日 | 2021-10-23 | | 公開日 | 2021-12-15 | | 最終更新日 | 2022-01-26 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Structure and mechanism of the SGLT family of glucose transporters.
Nature, 601, 2022
|
|
7SL8
 
 | | CryoEM structure of SGLT1 at 3.4 A resolution | | 分子名称: | CHOLESTEROL, Sodium/glucose cotransporter 1, nanobody Nb1 | | 著者 | Qu, Q, Han, L, Panova, O, Feng, L, Skiniotis, G. | | 登録日 | 2021-10-23 | | 公開日 | 2021-12-15 | | 最終更新日 | 2022-01-26 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structure and mechanism of the SGLT family of glucose transporters.
Nature, 601, 2022
|
|
6DA6
 
 | | Crystal structure of the TtnD decarboxylase from the tautomycetin biosynthesis pathway of Streptomyces griseochromogenes, apo form at 2.6 A resolution (P212121) | | 分子名称: | GLYCEROL, MAGNESIUM ION, UNKNOWN LIGAND, ... | | 著者 | Han, L, Rudolf, J.D, Chang, C.-Y, Miller, M.D, Soman, J, Phillips Jr, G.N, Shen, B, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2018-05-01 | | 公開日 | 2018-10-03 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.59 Å) | | 主引用文献 | Biochemical and Structural Characterization of TtnD, a Prenylated FMN-Dependent Decarboxylase from the Tautomycetin Biosynthetic Pathway.
ACS Chem. Biol., 13, 2018
|
|
6DA7
 
 | | Crystal structure of the TtnD decarboxylase from the tautomycetin biosynthesis pathway of Streptomyces griseochromogenes with apo form at 1.83 A resolution (I222) | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, SODIUM ION, ... | | 著者 | Han, L, Rudolf, J.D, Chang, C.-Y, Miller, M.D, Soman, J, Phillips Jr, G.N, Shen, B, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2018-05-01 | | 公開日 | 2018-10-03 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.83 Å) | | 主引用文献 | Biochemical and Structural Characterization of TtnD, a Prenylated FMN-Dependent Decarboxylase from the Tautomycetin Biosynthetic Pathway.
ACS Chem. Biol., 13, 2018
|
|
6DA9
 
 | | Crystal structure of the TtnD decarboxylase from the tautomycetin biosynthesis pathway of Streptomyces griseochromogenes with FMN bound at 2.05 A resolution | | 分子名称: | FLAVIN MONONUCLEOTIDE, GLYCEROL, MANGANESE (II) ION, ... | | 著者 | Han, L, Rudolf, J.D, Chang, C.-Y, Miller, M.D, Soman, J, Xu, W, Phillips Jr, G.N, Shen, B, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2018-05-01 | | 公開日 | 2018-10-03 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Biochemical and Structural Characterization of TtnD, a Prenylated FMN-Dependent Decarboxylase from the Tautomycetin Biosynthetic Pathway.
ACS Chem. Biol., 13, 2018
|
|
5N5O
 
 | |
5NH0
 
 | |
9C49
 
 | |
6XMF
 
 | |
6XMG
 
 | | Cryo-EM structure of Cas12g ternary complex | | 分子名称: | CRISPR-Cas, RNA (130-MER), RNA (5'-R(P*UP*UP*AP*AP*UP*GP*CP*GP*GP*UP*AP*GP*UP*UP*UP*AP*UP*CP*AP*CP*AP*GP*UP*U)-3'), ... | | 著者 | Chang, L, Li, Z, Zhang, H. | | 登録日 | 2020-06-30 | | 公開日 | 2021-01-13 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (4.8 Å) | | 主引用文献 | Cryo-EM structure of the RNA-guided ribonuclease Cas12g.
Nat.Chem.Biol., 17, 2021
|
|
