7BTE
 
 | | Lifeact-F-actin complex | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | 著者 | Kumari, A, Ragunath, V.K, Sirajuddin, M. | | 登録日 | 2020-04-01 | | 公開日 | 2020-05-20 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | Structural insights into actin filament recognition by commonly used cellular actin markers.
Embo J., 39, 2020
|
|
6M5G
 
 | | F-actin-Utrophin complex | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | 著者 | Kumari, A, Ragunath, V.K, Sirajuddin, M. | | 登録日 | 2020-03-10 | | 公開日 | 2020-05-20 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structural insights into actin filament recognition by commonly used cellular actin markers.
Embo J., 39, 2020
|
|
7BTI
 
 | | Phalloidin bound F-actin complex | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | 著者 | Kumari, A, Ragunath, V.K, Sirajuddin, M. | | 登録日 | 2020-04-01 | | 公開日 | 2020-05-20 | | 最終更新日 | 2025-04-09 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structural insights into actin filament recognition by commonly used cellular actin markers.
Embo J., 39, 2020
|
|
7BT7
 
 | | F-actin-ADP complex structure | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | 著者 | Kumari, A, Ragunath, V.K, Sirajuddin, M. | | 登録日 | 2020-03-31 | | 公開日 | 2020-05-20 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structural insights into actin filament recognition by commonly used cellular actin markers.
Embo J., 39, 2020
|
|
7F8R
 
 | |
6FC5
 
 | |
8BEW
 
 | |
3IX0
 
 | |
5FCH
 
 | | Crystal Structure of Xaa-Pro dipeptidase from Xanthomonas campestris, phosphate and Zn bound | | 分子名称: | DI(HYDROXYETHYL)ETHER, GLY-GLY-GLY, GLYCEROL, ... | | 著者 | Kumar, A, Are, V, Ghosh, B, Jamdar, S, Makde, R.D. | | 登録日 | 2015-12-15 | | 公開日 | 2016-12-07 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Crystal structure and biochemical investigations reveal novel mode of substrate selectivity and illuminate substrate inhibition and allostericity in a subfamily of Xaa-Pro dipeptidases
Biochim. Biophys. Acta, 1865, 2017
|
|
5FCF
 
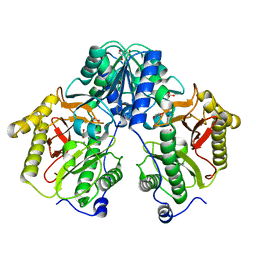 | | Crystal Structure of Xaa-Pro dipeptidase from Xanthomonas campestris, phosphate and Mn bound | | 分子名称: | DI(HYDROXYETHYL)ETHER, GLY-GLY-GLY, GLYCEROL, ... | | 著者 | Kumar, A, Are, V, Ghosh, B, Jamdar, S, Makde, R.D. | | 登録日 | 2015-12-15 | | 公開日 | 2016-12-07 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Crystal structure and biochemical investigations reveal novel mode of substrate selectivity and illuminate substrate inhibition and allostericity in a subfamily of Xaa-Pro dipeptidases.
Biochim. Biophys. Acta, 1865, 2017
|
|
7NPW
 
 | |
3MFY
 
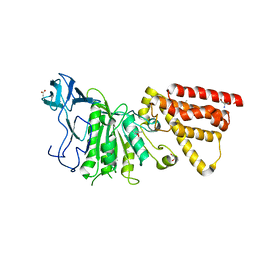 | | Structural characterization of the subunit A mutant F236A of the A-ATP synthase from Pyrococcus horikoshii | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETIC ACID, ... | | 著者 | Balakrishna, A.M, Kumar, A, Manimekali, M.S.S, Jeyakanthan, J, Gruber, G. | | 登録日 | 2010-04-05 | | 公開日 | 2010-07-07 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | The critical roles of residues P235 and F236 of subunit A of the motor protein A-ATP synthase in P-loop formation and nucleotide binding.
J.Mol.Biol., 401, 2010
|
|
3M4Y
 
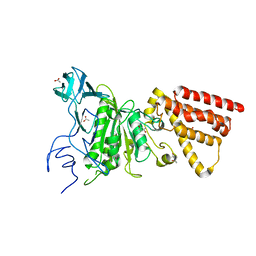 | | Structural characterization of the subunit A mutant P235A of the A-ATP synthase | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETIC ACID, ... | | 著者 | Manimekalai, M.S, Balakrishna, A.M, Kumar, A, Priya, R, Biukovic, G, Jeyakanthan, J, Gruber, G. | | 登録日 | 2010-03-12 | | 公開日 | 2011-01-26 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.38 Å) | | 主引用文献 | The critical roles of residues P235 and F236 of subunit A of the motor protein A-ATP synthase in P-loop formation and nucleotide binding.
J.Mol.Biol., 401, 2010
|
|
6FC6
 
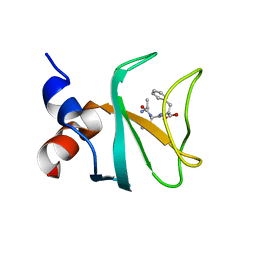 | |
7Q4W
 
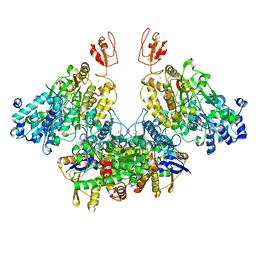 | | CryoEM structure of electron bifurcating Fe-Fe hydrogenase HydABC complex A. woodii in the oxidised state | | 分子名称: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, IRON/SULFUR CLUSTER, ... | | 著者 | Kumar, A, Saura, P, Poeverlein, M.C, Gamiz-Hernandez, A.P, Kaila, V.R.I, Mueller, V, Schuller, J.M. | | 登録日 | 2021-11-02 | | 公開日 | 2023-02-15 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (3.78 Å) | | 主引用文献 | Molecular Basis of the Electron Bifurcation Mechanism in the [FeFe]-Hydrogenase Complex HydABC.
J.Am.Chem.Soc., 145, 2023
|
|
3ND9
 
 | |
3ND8
 
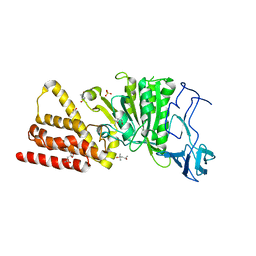 | | Structural characterization for the nucleotide binding ability of subunit A of the A1AO ATP synthase | | 分子名称: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, ACETIC ACID, ... | | 著者 | Kumar, A, Gruber, G. | | 登録日 | 2010-06-07 | | 公開日 | 2011-03-30 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | The transition-like state and Pi entrance into the catalytic a subunit of the biological engine A-ATP synthase.
J.Mol.Biol., 408, 2011
|
|
7VMF
 
 | |
8JCR
 
 | | Crystal structure of Calotropain FI from Calotropis gigantea (pH 6.0) | | 分子名称: | GLYCEROL, N-[N-[1-HYDROXYCARBOXYETHYL-CARBONYL]LEUCYLAMINO-BUTYL]-GUANIDINE, Procerain, ... | | 著者 | Kumar, A, Jamdar, S.N, Srivastava, G, Makde, R.D. | | 登録日 | 2023-05-11 | | 公開日 | 2024-05-22 | | 最終更新日 | 2025-04-23 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Crystal Structure of Papain-Like Cysteine Protease, Calotropain FI, Purified from the Latex of Calotropis gigantea.
J.Agric.Food Chem., 73, 2025
|
|
8JCQ
 
 | | Crystal structure of calotropain FI from Calotropis gigantea | | 分子名称: | N-[N-[1-HYDROXYCARBOXYETHYL-CARBONYL]LEUCYLAMINO-BUTYL]-GUANIDINE, Procerain | | 著者 | Kumar, A, Jamdar, S.N, Srivastava, G, Makde, R.D. | | 登録日 | 2023-05-11 | | 公開日 | 2024-05-22 | | 最終更新日 | 2025-04-23 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | Crystal Structure of Papain-Like Cysteine Protease, Calotropain FI, Purified from the Latex of Calotropis gigantea.
J.Agric.Food Chem., 73, 2025
|
|
7RBF
 
 | | Human DNA polymerase beta crosslinked binary complex - B | | 分子名称: | 1,2-ETHANEDIOL, 2-deoxy-3,5-di-O-phosphono-D-erythro-pentitol, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Reed, A.J, Kumar, A. | | 登録日 | 2021-07-06 | | 公開日 | 2022-03-09 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.84 Å) | | 主引用文献 | Interlocking activities of DNA polymerase beta in the base excision repair pathway.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7RBG
 
 | | Human DNA polymerase beta crosslinked ternary complex 1 | | 分子名称: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, 2-deoxy-3,5-di-O-phosphono-D-erythro-pentitol, CALCIUM ION, ... | | 著者 | Reed, A.J, Kumar, A. | | 登録日 | 2021-07-06 | | 公開日 | 2022-03-09 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Interlocking activities of DNA polymerase beta in the base excision repair pathway.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7RBE
 
 | | Human DNA polymerase beta crosslinked binary complex - A | | 分子名称: | 2-deoxy-3,5-di-O-phosphono-D-erythro-pentitol, CITRIC ACID, DNA (5'-D(*CP*CP*GP*AP*CP*GP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), ... | | 著者 | Reed, A.J, Kumar, A. | | 登録日 | 2021-07-06 | | 公開日 | 2022-03-09 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | Interlocking activities of DNA polymerase beta in the base excision repair pathway.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8A6T
 
 | | Cryo-EM structure of the electron bifurcating Fe-Fe hydrogenase HydABC complex from Thermoanaerobacter kivui in the reduced state | | 分子名称: | 2 IRON/2 SULFUR/5 CARBONYL/2 WATER INORGANIC CLUSTER, Electron bifurcating hydrogenase subunit HydA1, Electron bifurcating hydrogenase subunit HydB, ... | | 著者 | Kumar, A, Saura, P, Gamiz-Hernandez, A.P, Kaila, V.R.I, Mueller, V, Schuller, J.M. | | 登録日 | 2022-06-19 | | 公開日 | 2023-02-15 | | 最終更新日 | 2025-07-09 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Molecular Basis of the Electron Bifurcation Mechanism in the [FeFe]-Hydrogenase Complex HydABC.
J.Am.Chem.Soc., 145, 2023
|
|
8A5E
 
 | | Cryo-EM structure of the electron bifurcating Fe-Fe hydrogenase HydABC complex from Acetobacterium woodii in the reduced state | | 分子名称: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 2 IRON/2 SULFUR/5 CARBONYL/2 WATER INORGANIC CLUSTER, FE2/S2 (INORGANIC) CLUSTER, ... | | 著者 | Kumar, A, Saura, P, Gamiz-Hernandez, A.P, Kaila, V.R.I, Mueller, V, Schuller, J.M. | | 登録日 | 2022-06-14 | | 公開日 | 2023-02-22 | | 最終更新日 | 2023-03-29 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Molecular Basis of the Electron Bifurcation Mechanism in the [FeFe]-Hydrogenase Complex HydABC.
J.Am.Chem.Soc., 145, 2023
|
|
