1NU0
 
 | | Structure of the double mutant (L6M; F134M, SeMet form) of yqgF from Escherichia coli, a hypothetical protein | | 分子名称: | Hypothetical protein yqgF, SULFATE ION | | 著者 | Galkin, A, Sarikaya, E, Krajewski, W, Howard, A, Herzberg, O, Structure 2 Function Project (S2F) | | 登録日 | 2003-01-30 | | 公開日 | 2004-03-02 | | 最終更新日 | 2021-10-27 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structure of yqgF from Escherichia coli, a hypothetical protein
To be Published
|
|
1R8G
 
 | | Structure and function of YbdK | | 分子名称: | Hypothetical protein ybdK | | 著者 | Lehmann, C, Doseeva, V, Pullalarevu, S, Krajewski, W, Howard, A, Herzberg, O, Structure 2 Function Project (S2F) | | 登録日 | 2003-10-24 | | 公開日 | 2004-08-17 | | 最終更新日 | 2021-07-28 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | YbdK is a carboxylate-amine ligase with a gamma-glutamyl:Cysteine ligase activity: crystal structure and enzymatic assays
PROTEINS: STRUCT.,FUNCT.,GENET., 56, 2004
|
|
1L9Y
 
 | | FEZ-1-Y228A, A Mutant of the Metallo-beta-lactamase from Legionella gormanii | | 分子名称: | CHLORIDE ION, FEZ-1 b-lactamase, GLYCEROL, ... | | 著者 | Garcia-Saez, I, Mercuri, P.S, Galleni, M, Dideberg, O. | | 登録日 | 2002-03-27 | | 公開日 | 2003-07-01 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | Three-dimensional Structure of FEZ-1, a Monomeric Subclass B3 Metallo-beta-lactamase
from Fluoribacter gormanii, in Native Form and in Complex with -Captopril
J.Mol.Biol., 325, 2003
|
|
1PYM
 
 | |
1MQO
 
 | |
1LVG
 
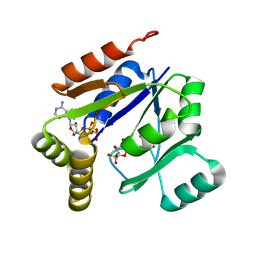 | | Crystal structure of mouse guanylate kinase in complex with GMP and ADP | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-MONOPHOSPHATE, Guanylate kinase, ... | | 著者 | Sekulic, N, Shuvalova, L, Spangenberg, O, Konrad, M, Lavie, A. | | 登録日 | 2002-05-28 | | 公開日 | 2002-12-11 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural characterization of the closed
conformation of mouse guanylate kinase.
J.Biol.Chem., 277, 2002
|
|
1NMN
 
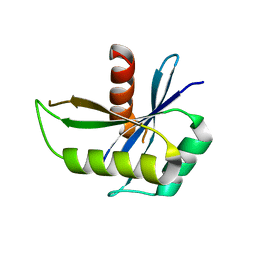 | | Structure of yqgF from Escherichia coli, a hypothetical protein | | 分子名称: | Hypothetical protein yqgF | | 著者 | Galkin, A, Sarikaya, E, Krajewski, W, Howard, A, Herzberg, O, Structure 2 Function Project (S2F) | | 登録日 | 2003-01-10 | | 公開日 | 2004-03-02 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure of yqgF from Escherichia coli, a hypothetical protein
To be Published
|
|
1SLT
 
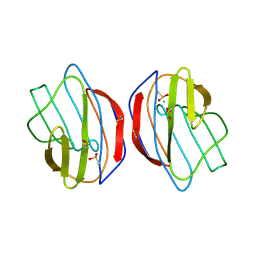 | |
1S2W
 
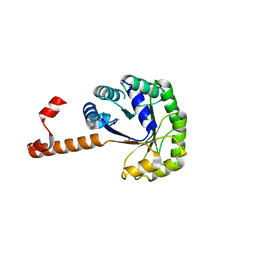 | | Crystal structure of phosphoenolpyruvate mutase in high ionic strength | | 分子名称: | Phosphoenolpyruvate phosphomutase, SULFATE ION | | 著者 | Liu, S, Lu, Z, Han, Y, Jia, Y, Howard, A, Dunaway-Mariano, D, Herzberg, O. | | 登録日 | 2004-01-11 | | 公開日 | 2004-05-04 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.69 Å) | | 主引用文献 | Conformational Flexibility of PEP Mutase
Biochemistry, 43, 2004
|
|
1KO2
 
 | | VIM-2, a Zn-beta-lactamase from Pseudomonas aeruginosa with an oxidized Cys (cysteinesulfonic) | | 分子名称: | ACETATE ION, VIM-2 metallo-beta-lactamase, ZINC ION | | 著者 | Garcia-Saez, I, Docquier, J.-D, Rossolini, G.M, Dideberg, O. | | 登録日 | 2001-12-20 | | 公開日 | 2003-09-02 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | The three-dimensional structure of VIM-2, a Zn-beta-lactamase from Pseudomonas aeruginosa in its reduced and oxidised form
J.Mol.Biol., 375, 2008
|
|
1NNX
 
 | | Structure of the hypothetical protein ygiW from E. coli. | | 分子名称: | Protein ygiW, SULFATE ION | | 著者 | Lehmann, C, Galkin, A, Pullalarevu, S, Sarikaya, E, Krajewski, W, Lim, K, Howard, A, Herzberg, O, Structure 2 Function Project (S2F) | | 登録日 | 2003-01-14 | | 公開日 | 2004-03-09 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Structure of the hypothetical protein ygiW from E. coli.
To be Published
|
|
1S2U
 
 | | Crystal structure of the D58A phosphoenolpyruvate mutase mutant protein | | 分子名称: | DI(HYDROXYETHYL)ETHER, Phosphoenolpyruvate phosphomutase | | 著者 | Liu, S, Lu, Z, Han, Y, Jia, Y, Howard, A, Dunaway-Mariano, D, Herzberg, O. | | 登録日 | 2004-01-11 | | 公開日 | 2004-05-04 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Conformational Flexibility of PEP Mutase
Biochemistry, 43, 2004
|
|
2UWX
 
 | | Active site restructuring regulates ligand recognition in class A penicillin-binding proteins | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, PENICILLIN-BINDING PROTEIN 1B, ... | | 著者 | Macheboeuf, P, DiGuilmi, A.M, Job, V, Vernet, T, Dideberg, O, Dessen, A. | | 登録日 | 2007-03-23 | | 公開日 | 2007-04-03 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.39 Å) | | 主引用文献 | Active Site Restructuring Regulates Ligand Recognition in Class a Penicillin-Binding Proteins
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
1MW5
 
 | | Structure of HI1480 from Haemophilus influenzae | | 分子名称: | HYPOTHETICAL PROTEIN HI1480 | | 著者 | Lim, K, Sarikaya, E, Howard, A, Galkin, A, Herzberg, O, Structure 2 Function Project (S2F) | | 登録日 | 2002-09-27 | | 公開日 | 2003-11-18 | | 最終更新日 | 2017-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Novel structure and nucleotide binding properties of HI1480 from Haemophilus influenzae: a protein with no known sequence homologues
PROTEINS: STRUCT.,FUNCT.,GENET., 56, 2004
|
|
1RW1
 
 | | YFFB (PA3664) PROTEIN | | 分子名称: | ISOPROPYL ALCOHOL, conserved hypothetical protein yffB | | 著者 | Teplyakov, A, Pullalarevu, S, Obmolova, G, Doseeva, V, Galkin, A, Herzberg, O, Dauter, M, Dauter, Z, Gilliland, G.L, Structure 2 Function Project (S2F) | | 登録日 | 2003-12-15 | | 公開日 | 2004-11-02 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.02 Å) | | 主引用文献 | Crystal structure of the YffB protein from Pseudomonas aeruginosa suggests a glutathione-dependent thiol reductase function.
Bmc Struct.Biol., 4, 2004
|
|
1MWQ
 
 | | Structure of HI0828, a Hypothetical Protein from Haemophilus influenzae with a Putative Active-Site Phosphohistidine | | 分子名称: | CACODYLATE ION, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Willis, M.A, Krajewski, W, Chalamasetty, V.R, Reddy, P, Howard, A, Herzberg, O, Structure 2 Function Project (S2F) | | 登録日 | 2002-09-30 | | 公開日 | 2003-11-25 | | 最終更新日 | 2019-07-24 | | 実験手法 | X-RAY DIFFRACTION (0.99 Å) | | 主引用文献 | Structure of YciI from Haemophilus influenzae (HI0828) reveals a ferredoxin-like alpha/beta-fold with a histidine/aspartate centered catalytic site
Proteins, 59, 2005
|
|
1MWW
 
 | | THE STRUCTURE OF THE HYPOTHETICAL PROTEIN HI1388.1 FROM HAEMOPHILUS INFLUENZAE REVEALS A TAUTOMERASE/MIF FOLD | | 分子名称: | CHLORIDE ION, GLUTAMIC ACID, HYPOTHETICAL PROTEIN HI1388.1 | | 著者 | Lehmann, C, Pullalarevu, S, Krajewski, W, Galkin, A, Howard, A, Herzberg, O, Structure 2 Function Project (S2F) | | 登録日 | 2002-10-01 | | 公開日 | 2003-11-18 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.08 Å) | | 主引用文献 | Structure of the Hypothetical Protein HI1388.1 from Haemophilus influenzae
To be Published
|
|
1MXI
 
 | | Structure of YibK from Haemophilus influenzae (HI0766): a Methyltransferase with a Cofactor Bound at a Site Formed by a Knot | | 分子名称: | Hypothetical tRNA/rRNA methyltransferase HI0766, IODIDE ION, S-ADENOSYL-L-HOMOCYSTEINE | | 著者 | Lim, K, Zhang, H, Tempczyk, A, Bonander, N, Toedt, J, Howard, A, Eisenstein, E, Herzberg, O, Structure 2 Function Project (S2F) | | 登録日 | 2002-10-02 | | 公開日 | 2003-02-25 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structure of the YibK methyltransferase from Haemophilus influenzae (HI0766): a Cofactor Bound at a Site Formed by a Knot
Proteins, 51, 2003
|
|
1M1B
 
 | | Crystal Structure of Phosphoenolpyruvate Mutase Complexed with Sulfopyruvate | | 分子名称: | MAGNESIUM ION, PHOSPHOENOLPYRUVATE PHOSPHOMUTASE, SULFOPYRUVATE | | 著者 | Liu, S, Lu, Z, Jia, Y, Dunaway-Mariano, D, Herzberg, O. | | 登録日 | 2002-06-18 | | 公開日 | 2002-08-28 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Dissociative phosphoryl transfer in PEP mutase catalysis: structure of the enzyme/sulfopyruvate complex and kinetic properties of mutants.
Biochemistry, 41, 2002
|
|
1M32
 
 | | Crystal Structure of 2-aminoethylphosphonate Transaminase | | 分子名称: | 2-aminoethylphosphonate-pyruvate aminotransferase, PHOSPHATE ION, PHOSPHONOACETALDEHYDE, ... | | 著者 | Chen, C.C.H, Zhang, H, Kim, A.D, Howard, A, Sheldrick, G.M, Mariano-Dunnaway, D, Herzberg, O. | | 登録日 | 2002-06-26 | | 公開日 | 2002-11-20 | | 最終更新日 | 2017-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Degradation Pathway of the Phosphonate Ciliatine: Crystal Structure of 2-Aminoethylphosphonate Transaminase
Biochemistry, 41, 2002
|
|
1RXX
 
 | | Structure of arginine deiminase | | 分子名称: | Arginine deiminase | | 著者 | Galkin, A, Kulakova, L, Sarikaya, E, Lim, K, Howard, A, Herzberg, O, Structure 2 Function Project (S2F) | | 登録日 | 2003-12-18 | | 公開日 | 2004-01-13 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Structural insight into arginine degradation by arginine deiminase, an antibacterial and parasite drug target.
J.Biol.Chem., 279, 2004
|
|
1S2V
 
 | | Crystal structure of phosphoenolpyruvate mutase complexed with Mg(II) | | 分子名称: | MAGNESIUM ION, Phosphoenolpyruvate phosphomutase | | 著者 | Liu, S, Lu, Z, Han, Y, Jia, Y, Howard, A, Dunaway-Mariano, D, Herzberg, O. | | 登録日 | 2004-01-11 | | 公開日 | 2004-05-04 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Conformational Flexibility of PEP Mutase
Biochemistry, 43, 2004
|
|
1NO5
 
 | | Structure of HI0073 from Haemophilus influenzae, the nucleotide binding domain of the HI0073/HI0074 two protein nucleotidyl transferase. | | 分子名称: | GLYCEROL, Hypothetical protein HI0073, SODIUM ION, ... | | 著者 | Lehmann, C, Pullalarevu, S, Galkin, A, Krajewski, W, Willis, M.A, Howard, A, Herzberg, O, Structure 2 Function Project (S2F) | | 登録日 | 2003-01-15 | | 公開日 | 2004-03-16 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structure of HI0073 from Haemophilus influenzae, the nucleotide-binding domain of a two-protein nucleotidyl transferase
Proteins, 60, 2005
|
|
1TFF
 
 | | Structure of Otubain-2 | | 分子名称: | Ubiquitin thiolesterase protein OTUB2 | | 著者 | Nanao, M.H, Tcherniuk, S.O, Chroboczek, J, Dideberg, O, Dessen, A, Balakirev, M.Y. | | 登録日 | 2004-05-27 | | 公開日 | 2004-08-03 | | 最終更新日 | 2021-10-27 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal structure of human otubain 2.
Embo Rep., 5, 2004
|
|
1ORT
 
 | | ORNITHINE TRANSCARBAMOYLASE FROM PSEUDOMONAS AERUGINOSA | | 分子名称: | ORNITHINE TRANSCARBAMOYLASE | | 著者 | Villeret, V, Dideberg, O. | | 登録日 | 1995-08-24 | | 公開日 | 1996-12-07 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Crystal structure of Pseudomonas aeruginosa catabolic ornithine transcarbamoylase at 3.0-A resolution: a different oligomeric organization in the transcarbamoylase family.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
