6MBG
 
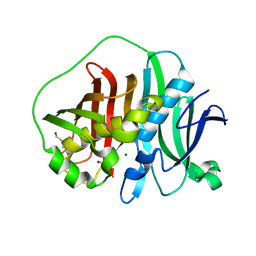 | |
6MBF
 
 | | GphF Dehydratase 1 | | 分子名称: | GphF Dehydratase 1, MAGNESIUM ION | | 著者 | Dodge, G.J, Smith, J.L. | | 登録日 | 2018-08-29 | | 公開日 | 2018-09-19 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.543 Å) | | 主引用文献 | Molecular Basis for Olefin Rearrangement in the Gephyronic Acid Polyketide Synthase.
ACS Chem. Biol., 13, 2018
|
|
6NEV
 
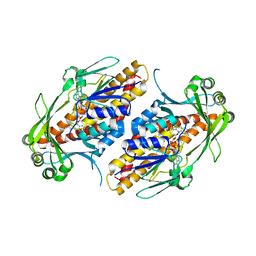 | | FAD-dependent monooxygenase TropB from T. stipitatus Y239F Variant | | 分子名称: | CHLORIDE ION, FAD-dependent monooxygenase tropB, FLAVIN-ADENINE DINUCLEOTIDE | | 著者 | Rodriguez Benitez, A, Tweedy, S.E, Baker Dockrey, S.A, Lukowski, A.L, Wymore, T, Khare, D, Palfey, B.A, Smith, J.L, Narayan, A.R.H. | | 登録日 | 2018-12-18 | | 公開日 | 2019-08-14 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.303 Å) | | 主引用文献 | Structural basis for selectivity in flavin-dependent monooxygenase-catalyzed oxidative dearomatization.
Acs Catalysis, 9, 2019
|
|
4O6C
 
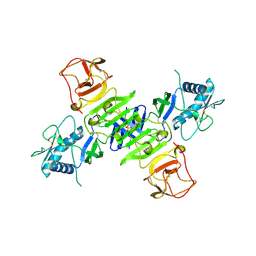 | |
1OX6
 
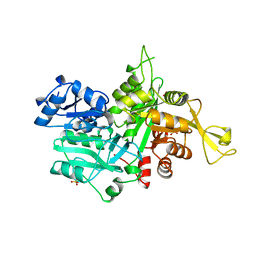 | |
1OX4
 
 | |
1OX5
 
 | |
4MYY
 
 | |
4MYZ
 
 | |
7UCI
 
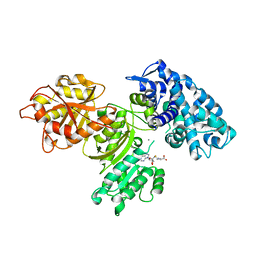 | | SxtA Methyltransferase and decarboxylase didomain in complex with Mn2+ and SAH | | 分子名称: | GLYCEROL, MANGANESE (II) ION, Polyketide synthase-related protein, ... | | 著者 | Lao, Y, Skiba, M.A, Smith, J.L. | | 登録日 | 2022-03-16 | | 公開日 | 2022-06-01 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structural Basis for Control of Methylation Extent in Polyketide Synthase Metal-Dependent C -Methyltransferases.
Acs Chem.Biol., 17, 2022
|
|
7UCL
 
 | |
4MZ0
 
 | |
4GBM
 
 | |
4H5L
 
 | |
4H5Q
 
 | |
6AL7
 
 | | Crystal structure HpiC1 F138S | | 分子名称: | 12-epi-hapalindole C/U synthase, CALCIUM ION | | 著者 | Newmister, S.A, Li, S, Garcia-Borras, M, Sanders, J.N, Yang, S, Lowell, A.N, Yu, F, Smith, J.L, Williams, R.M, Houk, K.N, Sherman, D.H. | | 登録日 | 2017-08-07 | | 公開日 | 2018-03-07 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.687 Å) | | 主引用文献 | Structural basis of the Cope rearrangement and cyclization in hapalindole biogenesis.
Nat. Chem. Biol., 14, 2018
|
|
6AL6
 
 | | Crystal structure HpiC1 in P42 space group | | 分子名称: | 12-epi-hapalindole C/U synthase, CALCIUM ION | | 著者 | Newmister, S.A, Li, S, Garcia-Borras, M, Sanders, J.N, Yang, S, Lowell, A.N, Yu, F, Smith, J.L, Williams, R.M, Houk, K.N, Sherman, D.H. | | 登録日 | 2017-08-07 | | 公開日 | 2018-03-07 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.088 Å) | | 主引用文献 | Structural basis of the Cope rearrangement and cyclization in hapalindole biogenesis.
Nat. Chem. Biol., 14, 2018
|
|
6AL8
 
 | | Crystal structure HpiC1 Y101F/F138S | | 分子名称: | 1,2-ETHANEDIOL, 12-epi-hapalindole C/U synthase, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | 著者 | Newmister, S.A, Li, S, Garcia-Borras, M, Sanders, J.N, Yang, S, Lowell, A.N, Yu, F, Smith, J.L, Williams, R.M, Houk, K.N, Sherman, D.H. | | 登録日 | 2017-08-07 | | 公開日 | 2018-03-07 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.641 Å) | | 主引用文献 | Structural basis of the Cope rearrangement and cyclization in hapalindole biogenesis.
Nat. Chem. Biol., 14, 2018
|
|
6BEV
 
 | |
6B3A
 
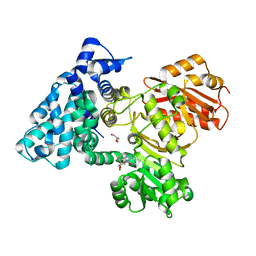 | |
6B3B
 
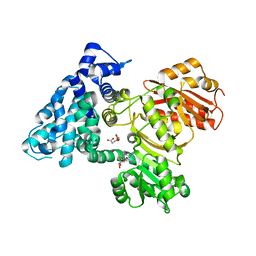 | | AprA Methyltransferase 1 - GNAT in complex with Mn2+ , SAM, and Malonate | | 分子名称: | AprA Methyltransferase 1, GLYCEROL, MALONATE ION, ... | | 著者 | Skiba, M.A, Smith, J.L. | | 登録日 | 2017-09-21 | | 公開日 | 2017-11-15 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | A Mononuclear Iron-Dependent Methyltransferase Catalyzes Initial Steps in Assembly of the Apratoxin A Polyketide Starter Unit.
ACS Chem. Biol., 12, 2017
|
|
6B39
 
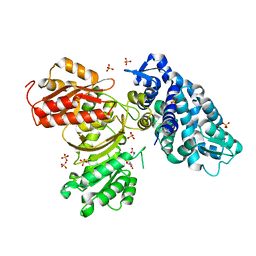 | | AprA Methyltransferase 1 - GNAT in complex with SAH | | 分子名称: | AprA Methyltransferase 1, S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION | | 著者 | Skiba, M.A, Smith, J.L. | | 登録日 | 2017-09-21 | | 公開日 | 2017-11-15 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.392 Å) | | 主引用文献 | A Mononuclear Iron-Dependent Methyltransferase Catalyzes Initial Steps in Assembly of the Apratoxin A Polyketide Starter Unit.
ACS Chem. Biol., 12, 2017
|
|
4GOX
 
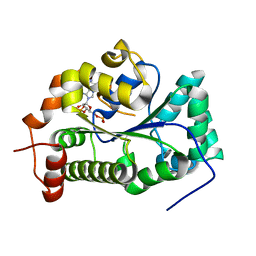 | |
4H5O
 
 | |
5DOZ
 
 | |
