5DOV
 
 | |
4X7Z
 
 | | MycF mycinamicin III 3'-O-methyltransferase (E35Q, M56A, E139A variant) in complex with Mg, SAH and mycinamicin III (substrate) | | 分子名称: | DIMETHYL SULFOXIDE, MAGNESIUM ION, MYCINAMICIN III, ... | | 著者 | Bernard, S.M, Smith, J.L. | | 登録日 | 2014-12-09 | | 公開日 | 2015-03-04 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.44 Å) | | 主引用文献 | Structural Basis of Substrate Specificity and Regiochemistry in the MycF/TylF Family of Sugar O-Methyltransferases.
Acs Chem.Biol., 10, 2015
|
|
4X7X
 
 | | MycF mycinamicin III 3'-O-methyltransferase (E35Q, E139A variant) in complex with Mg, SAH and macrocin | | 分子名称: | 2-[(4R,5S,6S,7R,9R,11E,13E,15R,16R)-6-[(2R,3R,4R,5S,6R)-4-(dimethylamino)-5-[(2S,4R,5S,6S)-4,6-dimethyl-4,5-bis(oxidanyl)oxan-2-yl]oxy-6-methyl-3-oxidanyl-oxan-2-yl]oxy-16-ethyl-15-[[(2R,3R,4R,5S,6R)-3-methoxy-6-methyl-4,5-bis(oxidanyl)oxan-2-yl]oxymethyl]-5,9,13-trimethyl-4-oxidanyl-2,10-bis(oxidanylidene)-1-oxacyclohexadeca-11,13-dien-7-yl]ethanal, MAGNESIUM ION, Mycinamicin III 3''-O-methyltransferase, ... | | 著者 | Bernard, S.M, Smith, J.L. | | 登録日 | 2014-12-09 | | 公開日 | 2015-03-11 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structural Basis of Substrate Specificity and Regiochemistry in the MycF/TylF Family of Sugar O-Methyltransferases.
Acs Chem.Biol., 10, 2015
|
|
4X7Y
 
 | |
4X7V
 
 | | MycF mycinamicin III 3'-O-methyltransferase (E35Q, E139A variant) in complex with Mg, SAH and mycinamicin IV (product) | | 分子名称: | MAGNESIUM ION, MYCINAMICIN IV, Mycinamicin III 3''-O-methyltransferase, ... | | 著者 | Bernard, S.M, Smith, J.L. | | 登録日 | 2014-12-09 | | 公開日 | 2015-03-04 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Structural Basis of Substrate Specificity and Regiochemistry in the MycF/TylF Family of Sugar O-Methyltransferases.
Acs Chem.Biol., 10, 2015
|
|
5DOZ
 
 | |
5DP1
 
 | |
5DP2
 
 | |
5KP8
 
 | |
5KP5
 
 | |
5KP7
 
 | |
7R7G
 
 | |
7R7F
 
 | |
7R7E
 
 | |
5KP6
 
 | |
3NNL
 
 | |
3NNF
 
 | | Halogenase domain from CurA module with Fe, chloride, and alpha-ketoglutarate | | 分子名称: | 2-OXOGLUTARIC ACID, CHLORIDE ION, CurA, ... | | 著者 | Khare, D, Smith, J.L. | | 登録日 | 2010-06-23 | | 公開日 | 2010-07-28 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.201 Å) | | 主引用文献 | Conformational switch triggered by alpha-ketoglutarate in a halogenase of curacin A biosynthesis
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3NNM
 
 | |
3NNJ
 
 | |
2Q34
 
 | |
2Q35
 
 | |
2Q2X
 
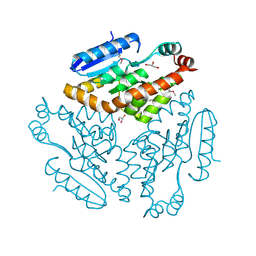 | |
3GWZ
 
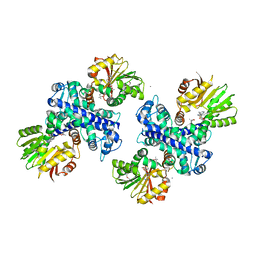 | | Structure of the Mitomycin 7-O-methyltransferase MmcR | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, MmcR, ... | | 著者 | Singh, S, Chang, A, Bingman, C.A, Phillips Jr, G.N, Thorson, J.S. | | 登録日 | 2009-04-01 | | 公開日 | 2010-04-07 | | 最終更新日 | 2017-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.91 Å) | | 主引用文献 | Structural characterization of the mitomycin 7-O-methyltransferase.
Proteins, 79, 2011
|
|
3GXO
 
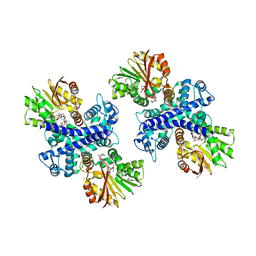 | | Structure of the Mitomycin 7-O-methyltransferase MmcR with bound Mitomycin A | | 分子名称: | CALCIUM ION, MmcR, S-ADENOSYL-L-HOMOCYSTEINE, ... | | 著者 | Singh, S, Chang, A, Bingman, C.A, Phillips Jr, G.N, Thorson, J.S. | | 登録日 | 2009-04-02 | | 公開日 | 2010-04-21 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural characterization of the mitomycin 7-O-methyltransferase.
Proteins, 79, 2011
|
|
1KMZ
 
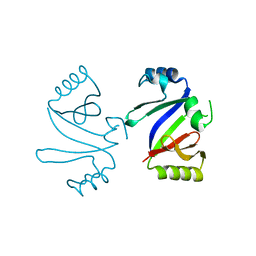 | | MOLECULAR BASIS OF MITOMYCIN C RESICTANCE IN STREPTOMYCES: CRYSTAL STRUCTURES OF THE MRD PROTEIN WITH AND WITHOUT A DRUG DERIVATIVE | | 分子名称: | mitomycin-binding protein | | 著者 | Martin, T.W, Dauter, Z, Devedjiev, Y, Sheffield, P, Jelen, F, He, M, Sherman, D, Otlewski, J, Derewenda, Z.S, Derewenda, U. | | 登録日 | 2001-12-17 | | 公開日 | 2002-07-19 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Molecular basis of mitomycin C resistance in streptomyces: structure and function of the MRD protein.
Structure, 10, 2002
|
|
