3AHN
 
 | | PZ PEPTIDASE A with Inhibitor 1 | | 分子名称: | ACETATE ION, N~2~-{(2S)-3-[(R)-hydroxy{(1R)-2-phenyl-1-[(phenylacetyl)amino]ethyl}phosphoryl]-2-methylpropanoyl}-L-lysyl-D-serine, Oligopeptidase, ... | | 著者 | Nakano, H. | | 登録日 | 2010-04-25 | | 公開日 | 2010-08-25 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | The exquisite structure and reaction mechanism of bacterial Pz-peptidase A toward collagenous peptides: X-ray crystallographic structure analysis of PZ-peptidase a reveals differences from mammalian thimet oligopeptidase.
J.Biol.Chem., 285, 2010
|
|
3WQ4
 
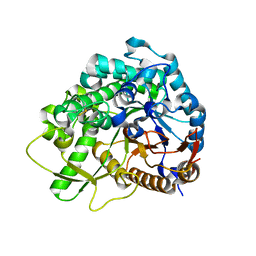 | | Crystal structure of beta-primeverosidase | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-primeverosidase | | 著者 | Saino, H. | | 登録日 | 2014-01-22 | | 公開日 | 2014-04-23 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structures of beta-primeverosidase in complex with disaccharide amidine inhibitors.
J.Biol.Chem., 289, 2014
|
|
3WQ5
 
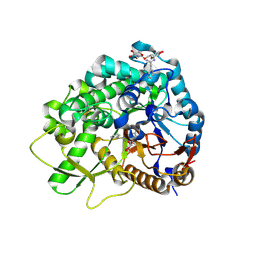 | |
2EII
 
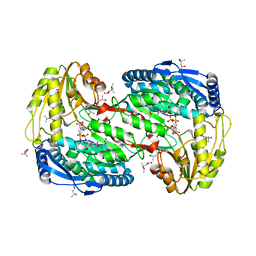 | | Crystal analysis of delta1-pyrroline-5-carboxylate dehydrogenase from Thermus thermophilus with bound L-valine and NAD. | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1-pyrroline-5-carboxylate dehydrogenase, ACETATE ION, ... | | 著者 | Inagaki, E, Sakamoto, K, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2007-03-13 | | 公開日 | 2007-09-18 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.88 Å) | | 主引用文献 | Crystal structure analysis of delta1-pyrroline-5-carboxylate dehydrogenase in ternary complex with inhibitor and NAD.
To be Published
|
|
2EHQ
 
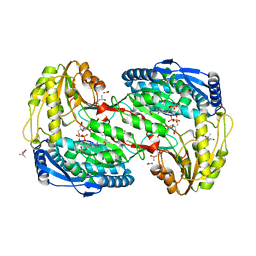 | | Crystal analysis of 1-pyrroline-5-carboxylate dehydrogenase from thermus with bound NADP | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1-pyrroline-5-carboxylate dehydrogenase, ACETATE ION, ... | | 著者 | Inagaki, E, Sakamoto, K, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2007-03-07 | | 公開日 | 2007-05-01 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | New insights into the binding mode of coenzymes: structure of Thermus thermophilus Delta1-pyrroline-5-carboxylate dehydrogenase complexed with NADP+.
Acta Crystallogr.,Sect.F, 63, 2007
|
|
3B1D
 
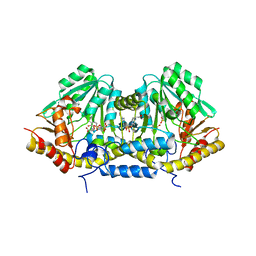 | | Crystal structure of betaC-S lyase from Streptococcus anginosus in complex with L-serine: External aldimine form | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, BetaC-S lyase, PYRIDOXAL-5'-PHOSPHATE, ... | | 著者 | Kezuka, Y, Yoshida, Y, Nonaka, T. | | 登録日 | 2011-06-29 | | 公開日 | 2012-06-27 | | 最終更新日 | 2025-03-26 | | 実験手法 | X-RAY DIFFRACTION (1.66 Å) | | 主引用文献 | Structural insights into catalysis by beta C-S lyase from Streptococcus anginosus
Proteins, 80, 2012
|
|
3B1C
 
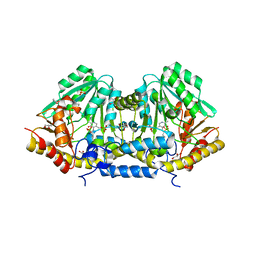 | | Crystal structure of betaC-S lyase from Streptococcus anginosus: Internal aldimine form | | 分子名称: | BetaC-S lyase, GLYCEROL, PYRIDOXAL-5'-PHOSPHATE, ... | | 著者 | Kezuka, Y, Yoshida, Y, Nonaka, T. | | 登録日 | 2011-06-29 | | 公開日 | 2012-06-27 | | 最終更新日 | 2025-03-26 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | Structural insights into catalysis by beta C-S lyase from Streptococcus anginosus
Proteins, 80, 2012
|
|
3B1E
 
 | | Crystal structure of betaC-S lyase from Streptococcus anginosus in complex with L-serine: alpha-Aminoacrylate form | | 分子名称: | 2-{[(E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}prop-2-enoic acid, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETATE ION, ... | | 著者 | Kezuka, Y, Yoshida, Y, Nonaka, T. | | 登録日 | 2011-06-29 | | 公開日 | 2012-06-27 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | Structural insights into catalysis by beta C-S lyase from Streptococcus anginosus
Proteins, 80, 2012
|
|
1ITX
 
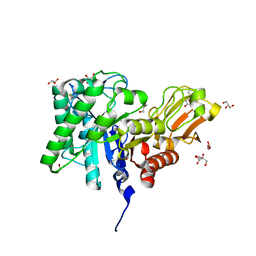 | | Catalytic Domain of Chitinase A1 from Bacillus circulans WL-12 | | 分子名称: | GLYCEROL, Glycosyl Hydrolase | | 著者 | Iwahori, F, Matsumoto, T, Watanabe, T, Nonaka, T. | | 登録日 | 2002-02-13 | | 公開日 | 2002-03-13 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Three-dimensional structure of the catalytic domain of chitinase A1 from Bacillus circulans WL-12 at a very high resolution
PROC.JPN.ACAD.,SER.B, 75, 1999
|
|
3VX8
 
 | |
5JH9
 
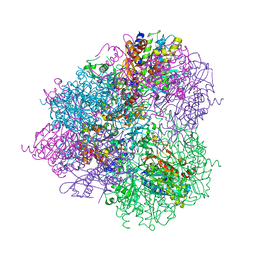 | | Crystal structure of prApe1 | | 分子名称: | CACODYLATE ION, Vacuolar aminopeptidase 1, ZINC ION | | 著者 | Noda, N.N, Adachi, W, Inagaki, F. | | 登録日 | 2016-04-20 | | 公開日 | 2016-06-29 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural Basis for Receptor-Mediated Selective Autophagy of Aminopeptidase I Aggregates
Cell Rep, 16, 2016
|
|
5JGE
 
 | |
5JGF
 
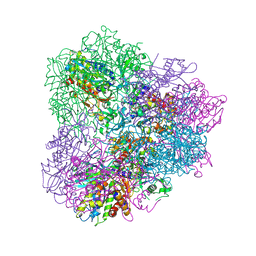 | | Crystal structure of mApe1 | | 分子名称: | Vacuolar aminopeptidase 1, ZINC ION | | 著者 | Noda, N.N, Adachi, W, Inagaki, F. | | 登録日 | 2016-04-20 | | 公開日 | 2016-06-29 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.83 Å) | | 主引用文献 | Structural Basis for Receptor-Mediated Selective Autophagy of Aminopeptidase I Aggregates
Cell Rep, 16, 2016
|
|
5JHC
 
 | |
6AAG
 
 | |
6AAF
 
 | |
6A9E
 
 | |
6A9J
 
 | |
2ZPN
 
 | |
2JPH
 
 | |
7BRU
 
 | |
7BRN
 
 | | Crystal structure of Atg40 AIM fused to Atg8 | | 分子名称: | 1,2-ETHANEDIOL, Autophagy-related protein 40,Autophagy-related protein 8, L-EPINEPHRINE | | 著者 | Yamasaki, A, Noda, N.N. | | 登録日 | 2020-03-29 | | 公開日 | 2020-07-08 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.231 Å) | | 主引用文献 | Super-assembly of ER-phagy receptor Atg40 induces local ER remodeling at contacts with forming autophagosomal membranes.
Nat Commun, 11, 2020
|
|
7BRQ
 
 | |
7BRT
 
 | | Crystal structure of Sec62 LIR fused to GABARAP | | 分子名称: | Translocation protein SEC62,Gamma-aminobutyric acid receptor-associated protein | | 著者 | Yamasaki, A, Noda, N.N. | | 登録日 | 2020-03-30 | | 公開日 | 2020-07-08 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.999 Å) | | 主引用文献 | Super-assembly of ER-phagy receptor Atg40 induces local ER remodeling at contacts with forming autophagosomal membranes.
Nat Commun, 11, 2020
|
|
3VH3
 
 | | Crystal structure of Atg7CTD-Atg8 complex | | 分子名称: | Autophagy-related protein 8, Ubiquitin-like modifier-activating enzyme ATG7, ZINC ION | | 著者 | Noda, N.N, Satoo, K, Inagaki, F. | | 登録日 | 2011-08-23 | | 公開日 | 2011-09-21 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural basis of Atg8 activation by a homodimeric E1, Atg7.
Mol.Cell, 44, 2011
|
|
