5YPZ
 
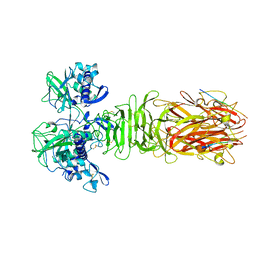 | | Crystal structure of minor pilin CofB from CFA/III complexed with N-terminal peptide fragment of CofJ | | 分子名称: | CofB, CofJ | | 著者 | Oki, H, Kawahara, K, Maruno, T, Imai, T, Muroga, Y, Fukakusa, S, Iwashita, T, Kobayashi, Y, Matsuda, S, Kodama, T, Iida, T, Yoshida, T, Ohkubo, T, Nakamura, S. | | 登録日 | 2017-11-04 | | 公開日 | 2018-06-27 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.521 Å) | | 主引用文献 | Interplay of a secreted protein with type IVb pilus for efficient enterotoxigenicEscherichia colicolonization.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7VNN
 
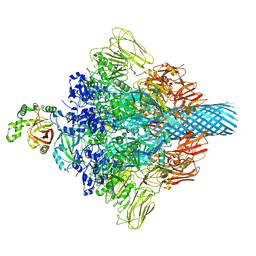 | | Complex structure of Clostridioides difficile enzymatic component (CDTa) and binding component (CDTb) pore with long stem | | 分子名称: | ADP-ribosylating binary toxin binding subunit CdtB, CALCIUM ION, CdtA | | 著者 | Yamada, T, Kawamoto, A, Yoshida, T, Sato, Y, Kato, T, Tsuge, H. | | 登録日 | 2021-10-11 | | 公開日 | 2022-10-26 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (2.64 Å) | | 主引用文献 | Cryo-EM structures of the translocational binary toxin complex CDTa-bound CDTb-pore from Clostridioides difficile.
Nat Commun, 13, 2022
|
|
7VNJ
 
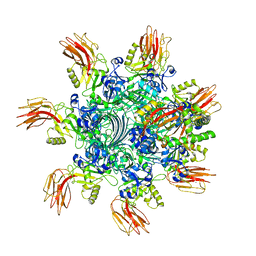 | | Complex structure of Clostridioides difficile enzymatic component (CDTa) and binding component (CDTb) pore with short stem | | 分子名称: | ADP-ribosylating binary toxin binding subunit CdtB, ADP-ribosyltransferase enzymatic component, CALCIUM ION | | 著者 | Yamada, T, Kawamoto, A, Yoshida, T, Sato, Y, Kato, T, Tsuge, H. | | 登録日 | 2021-10-11 | | 公開日 | 2022-10-26 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (2.56 Å) | | 主引用文献 | Cryo-EM structures of the translocational binary toxin complex CDTa-bound CDTb-pore from Clostridioides difficile.
Nat Commun, 13, 2022
|
|
7YVQ
 
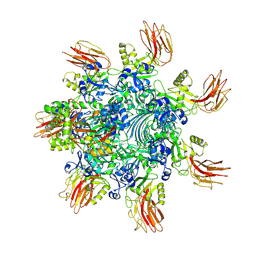 | | Complex structure of Clostridioides difficile binary toxin folded CDTa-bound CDTb-pore (short). | | 分子名称: | ADP-ribosylating binary toxin binding subunit CdtB, ADP-ribosylating binary toxin enzymatic subunit CdtA, CALCIUM ION | | 著者 | Yamada, T, Kawamoto, A, Yoshida, T, Sato, Y, Kato, T, Tsuge, H. | | 登録日 | 2022-08-19 | | 公開日 | 2022-10-26 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3.18 Å) | | 主引用文献 | Cryo-EM structures of the translocational binary toxin complex CDTa-bound CDTb-pore from Clostridioides difficile.
Nat Commun, 13, 2022
|
|
7YVS
 
 | | Complex structure of Clostridioides difficile binary toxin unfolded CDTa-bound CDTb-pore (short). | | 分子名称: | ADP-ribosylating binary toxin binding subunit CdtB, ADP-ribosylating binary toxin enzymatic subunit CdtA, CALCIUM ION | | 著者 | Yamada, T, Kawamoto, A, Yoshida, T, Sato, Y, Kato, T, Tsuge, H. | | 登録日 | 2022-08-19 | | 公開日 | 2022-10-26 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Cryo-EM structures of the translocational binary toxin complex CDTa-bound CDTb-pore from Clostridioides difficile.
Nat Commun, 13, 2022
|
|
1J1I
 
 | | Crystal structure of a His-tagged Serine Hydrolase Involved in the Carbazole Degradation (CarC enzyme) | | 分子名称: | meta cleavage compound hydrolase | | 著者 | Habe, H, Morii, K, Fushinobu, S, Nam, J.W, Ayabe, Y, Yoshida, T, Wakagi, T, Yamane, H, Nojiri, H, Omori, T. | | 登録日 | 2002-12-05 | | 公開日 | 2003-06-17 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.86 Å) | | 主引用文献 | Crystal structure of a histidine-tagged serine hydrolase involved in the carbazole degradation (CarC enzyme).
Biochem.Biophys.Res.Commun., 303, 2003
|
|
6JT5
 
 | | Crystal structure of PQQ doamin of Pyranose Dehydrogenase from Coprinopsis cinerea: apo-from | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Extracellular PQQ-dependent sugar dehydrogenase, ... | | 著者 | Takeda, K, Ishida, T, Yoshida, M, Samejima, M, Ohno, H, Igarashi, K, Nakamura, N. | | 登録日 | 2019-04-09 | | 公開日 | 2019-11-06 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Crystal Structure of the Catalytic and CytochromebDomains in a Eukaryotic Pyrroloquinoline Quinone-Dependent Dehydrogenase.
Appl.Environ.Microbiol., 85, 2019
|
|
7VNO
 
 | | Structure of aminotransferase | | 分子名称: | 1,2-ETHANEDIOL, 454aa long hypothetical 4-aminobutyrate aminotransferase, GLYCEROL, ... | | 著者 | Sakuraba, H, Ohshida, T, Ohshima, T. | | 登録日 | 2021-10-11 | | 公開日 | 2022-03-30 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of a novel type of ornithine delta-aminotransferase from the hyperthermophilic archaeon Pyrococcus horikoshii.
Int.J.Biol.Macromol., 208, 2022
|
|
7VO1
 
 | | Structure of aminotransferase-substrate complex | | 分子名称: | 454aa long hypothetical 4-aminobutyrate aminotransferase, N-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)-L-glutamic acid | | 著者 | Sakuraba, H, Ohshida, T, Ohshima, T. | | 登録日 | 2021-10-12 | | 公開日 | 2022-03-30 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.99 Å) | | 主引用文献 | Crystal structure of a novel type of ornithine delta-aminotransferase from the hyperthermophilic archaeon Pyrococcus horikoshii.
Int.J.Biol.Macromol., 208, 2022
|
|
7VNT
 
 | | Structure of aminotransferase-substrate complex | | 分子名称: | 1,2-ETHANEDIOL, 454aa long hypothetical 4-aminobutyrate aminotransferase, GLYCEROL, ... | | 著者 | Sakuraba, H, Ohshida, T, Ohshima, T. | | 登録日 | 2021-10-12 | | 公開日 | 2022-03-30 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | Crystal structure of a novel type of ornithine delta-aminotransferase from the hyperthermophilic archaeon Pyrococcus horikoshii.
Int.J.Biol.Macromol., 208, 2022
|
|
6JT6
 
 | | Crystal structure of cytochrome b domain of Pyranose Dehydrogenase from Coprinopsis cinerea | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | 著者 | Takeda, K, Ishida, T, Yoshida, M, Samejima, M, Ohno, H, Igarashi, K, Nakamura, N. | | 登録日 | 2019-04-09 | | 公開日 | 2019-11-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal Structure of the Catalytic and CytochromebDomains in a Eukaryotic Pyrroloquinoline Quinone-Dependent Dehydrogenase.
Appl.Environ.Microbiol., 85, 2019
|
|
1TAB
 
 | | STRUCTURE OF THE TRYPSIN-BINDING DOMAIN OF BOWMAN-BIRK TYPE PROTEASE INHIBITOR AND ITS INTERACTION WITH TRYPSIN | | 分子名称: | BOWMAN-BIRK TYPE PROTEINASE INHIBITOR, TRYPSIN | | 著者 | Tsunogae, Y, Tanaka, I, Yamane, T, Kikkawa, J.-I, Ashida, T, Ishikawa, C, Watanabe, K, Nakamura, S, Takahashi, K. | | 登録日 | 1990-10-15 | | 公開日 | 1992-01-15 | | 最終更新日 | 2024-06-05 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure of the trypsin-binding domain of Bowman-Birk type protease inhibitor and its interaction with trypsin.
J.Biochem.(Tokyo), 100, 1986
|
|
1JWQ
 
 | | Structure of the catalytic domain of CwlV, N-acetylmuramoyl-L-alanine amidase from Bacillus(Paenibacillus) polymyxa var.colistinus | | 分子名称: | N-ACETYLMURAMOYL-L-ALANINE AMIDASE CwlV, ZINC ION | | 著者 | Yamane, T, Koyama, Y, Nojiri, Y, Hikage, T, Akita, M, Suzuki, A, Shirai, T, Ise, F, Shida, T, Sekiguchi, J. | | 登録日 | 2001-09-05 | | 公開日 | 2003-11-18 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | The Structure of the catalytic domain of N-acetylmuramoyl-L-alanine amidase, a cell wall hydrolase from Bacillus polymyxa var.colistinus and its resemblance to the structure of carboxypeptidases
To be Published
|
|
7W05
 
 | | 12 mutant Ribonuclease from Hericium erinaceus GMP binding form | | 分子名称: | DI(HYDROXYETHYL)ETHER, GUANOSINE, Ribonuclease T1 | | 著者 | Takebe, K, Chida, T, Suzuki, M, Itagaki, T, Morita, Y, Uzawa, N, Kobayashi, H. | | 登録日 | 2021-11-17 | | 公開日 | 2022-11-23 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.29 Å) | | 主引用文献 | 12 mutant Ribonuclease from Hericium erinaceus GMP binding form
To Be Published
|
|
3MFP
 
 | | Atomic model of F-actin based on a 6.6 angstrom resolution cryoEM map | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle | | 著者 | Fujii, T, Iwane, A.H, Yanagida, T, Namba, K. | | 登録日 | 2010-04-03 | | 公開日 | 2010-09-29 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (6.6 Å) | | 主引用文献 | Direct visualization of secondary structures of F-actin by electron cryomicroscopy
Nature, 467, 2010
|
|
2KTD
 
 | | Solution structure of mouse lipocalin-type prostaglandin D synthase / substrate analog (U-46619) complex | | 分子名称: | (5Z)-7-{(1R,4S,5S,6R)-6-[(1E,3S)-3-hydroxyoct-1-en-1-yl]-2-oxabicyclo[2.2.1]hept-5-yl}hept-5-enoic acid, Prostaglandin-H2 D-isomerase | | 著者 | Shimamoto, S, Maruo, H, Yoshida, T, Kato, N, Ohkubo, T. | | 登録日 | 2010-01-27 | | 公開日 | 2011-02-02 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution Structure of Lipocalin-type Prostaglandin D synthase / Substrate analog complex reveals Open-Closed Conformational Change required for Substrate Recognition
To be Published
|
|
1CYC
 
 | | THE CRYSTAL STRUCTURE OF BONITO (KATSUO) FERROCYTOCHROME C AT 2.3 ANGSTROMS RESOLUTION. II. STRUCTURE AND FUNCTION | | 分子名称: | FERROCYTOCHROME C, HEME C | | 著者 | Tanaka, N, Yamane, T, Tsukihara, T, Ashida, T, Kakudo, M. | | 登録日 | 1976-08-01 | | 公開日 | 1976-10-06 | | 最終更新日 | 2021-03-03 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | The crystal structure of bonito (katsuo) ferrocytochrome c at 2.3 A resolution. II. Structure and function.
J.Biochem.(Tokyo), 77, 1975
|
|
5Y5F
 
 | | Structure of cytochrome P450nor in NO-bound state: damaged by low-dose (0.72 MGy) X-ray | | 分子名称: | GLYCEROL, NADP nitrous oxide-forming nitric oxide reductase, NITRIC OXIDE, ... | | 著者 | Tosha, T, Nomura, T, Nishida, T, Ueno, G, Murakami, H, Yamashita, K, Hirata, K, Yamamoto, M, Ago, H, Sugimoto, H, Shiro, Y, Kubo, M. | | 登録日 | 2017-08-09 | | 公開日 | 2017-12-06 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Capturing an initial intermediate during the P450nor enzymatic reaction using time-resolved XFEL crystallography and caged-substrate.
Nat Commun, 8, 2017
|
|
5Y5M
 
 | | SFX structure of cytochrome P450nor: a complete dark data without pump laser (resting state) | | 分子名称: | NADP nitrous oxide-forming nitric oxide reductase, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Tosha, T, Nomura, T, Nishida, T, Saeki, N, Okubayashi, K, Yamagiwa, R, Sugahara, M, Nakane, T, Yamashita, K, Hirata, K, Ueno, G, Kimura, T, Hisano, T, Muramoto, K, Sawai, H, Takeda, H, Mizohata, E, Yamashita, A, Kanematsu, Y, Takano, Y, Nango, E, Tanaka, R, Nureki, O, Ikemoto, Y, Murakami, H, Owada, S, Tono, K, Yabashi, M, Yamamoto, M, Ago, H, Iwata, S, Sugimoto, H, Shiro, Y, Kubo, M. | | 登録日 | 2017-08-09 | | 公開日 | 2018-08-08 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Capturing an initial intermediate during the P450nor enzymatic reaction using time-resolved XFEL crystallography and caged-substrate.
Nat Commun, 8, 2017
|
|
5Y5L
 
 | | Time-resolved SFX structure of cytochrome P450nor: dark-2 data in the absence of NADH (resting state) | | 分子名称: | NADP nitrous oxide-forming nitric oxide reductase, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Tosha, T, Nomura, T, Nishida, T, Saeki, N, Okubayashi, K, Yamagiwa, R, Sugahara, M, Nakane, T, Yamashita, K, Hirata, K, Ueno, G, Kimura, T, Hisano, T, Muramoto, K, Sawai, H, Takeda, H, Mizohata, E, Yamashita, A, Kanematsu, Y, Takano, Y, Nango, E, Tanaka, R, Nureki, O, Ikemoto, Y, Murakami, H, Owada, S, Tono, K, Yabashi, M, Yamamoto, M, Ago, H, Iwata, S, Sugimoto, H, Shiro, Y, Kubo, M. | | 登録日 | 2017-08-09 | | 公開日 | 2017-12-06 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Capturing an initial intermediate during the P450nor enzymatic reaction using time-resolved XFEL crystallography and caged-substrate.
Nat Commun, 8, 2017
|
|
5Y5H
 
 | | SF-ROX structure of cytochrome P450nor (NO-bound state) determined at SACLA | | 分子名称: | GLYCEROL, NADP nitrous oxide-forming nitric oxide reductase, NITRIC OXIDE, ... | | 著者 | Tosha, T, Nomura, T, Nishida, T, Yamagiwa, R, Yamashita, K, Hirata, K, Ueno, G, Kimura, T, Hisano, T, Muramoto, K, Sawai, H, Takeda, H, Yamashita, A, Murakami, H, Owada, S, Tono, K, Yabashi, M, Yamamoto, M, Ago, H, Sugimoto, H, Shiro, Y, Kubo, M. | | 登録日 | 2017-08-09 | | 公開日 | 2017-12-06 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Capturing an initial intermediate during the P450nor enzymatic reaction using time-resolved XFEL crystallography and caged-substrate.
Nat Commun, 8, 2017
|
|
5Y5I
 
 | | Time-resolved SFX structure of cytochrome P450nor: 20 ms after photo-irradiation of caged NO in the presence of NADH (NO-bound state), light data | | 分子名称: | GLYCEROL, NADP nitrous oxide-forming nitric oxide reductase, NITRIC OXIDE, ... | | 著者 | Tosha, T, Nomura, T, Nishida, T, Saeki, N, Okubayashi, K, Yamagiwa, R, Sugahara, M, Nakane, T, Yamashita, K, Hirata, K, Ueno, G, Kimura, T, Hisano, T, Muramoto, K, Sawai, H, Takeda, H, Mizohata, E, Yamashita, A, Kanematsu, Y, Takano, Y, Nango, E, Tanaka, R, Nureki, O, Ikemoto, Y, Murakami, H, Owada, S, Tono, K, Yabashi, M, Yamamoto, M, Ago, H, Iwata, S, Sugimoto, H, Shiro, Y, Kubo, M. | | 登録日 | 2017-08-09 | | 公開日 | 2017-12-06 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Capturing an initial intermediate during the P450nor enzymatic reaction using time-resolved XFEL crystallography and caged-substrate.
Nat Commun, 8, 2017
|
|
5Y5J
 
 | | Time-resolved SFX structure of cytochrome P450nor: dark-2 data in the presence of NADH (resting state) | | 分子名称: | GLYCEROL, NADP nitrous oxide-forming nitric oxide reductase, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Tosha, T, Nomura, T, Nishida, T, Saeki, N, Okubayashi, K, Yamagiwa, R, Sugahara, M, Nakane, T, Yamashita, K, Hirata, K, Ueno, G, Kimura, T, Hisano, T, Muramoto, K, Sawai, H, Takeda, H, Mizohata, E, Yamashita, A, Kanematsu, Y, Takano, Y, Nango, E, Tanaka, R, Nureki, O, Ikemoto, Y, Murakami, H, Owada, S, Tono, K, Yabashi, M, Yamamoto, M, Ago, H, Iwata, S, Sugimoto, H, Shiro, Y, Kubo, M. | | 登録日 | 2017-08-09 | | 公開日 | 2017-12-06 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Capturing an initial intermediate during the P450nor enzymatic reaction using time-resolved XFEL crystallography and caged-substrate.
Nat Commun, 8, 2017
|
|
5Y5K
 
 | | Time-resolved SFX structure of cytochrome P450nor : 20 ms after photo-irradiation of caged NO in the absence of NADH (NO-bound state), light data | | 分子名称: | NADP nitrous oxide-forming nitric oxide reductase, NITRIC OXIDE, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Tosha, T, Nomura, T, Nishida, T, Saeki, N, Okubayashi, K, Yamagiwa, R, Sugahara, M, Nakane, T, Yamashita, K, Hirata, K, Ueno, G, Kimura, T, Hisano, T, Muramoto, K, Sawai, H, Takeda, H, Mizohata, E, Yamashita, A, Kanematsu, Y, Takano, Y, Nango, E, Tanaka, R, Nureki, O, Ikemoto, Y, Murakami, H, Owada, S, Tono, K, Yabashi, M, Yamamoto, M, Ago, H, Iwata, S, Sugimoto, H, Shiro, Y, Kubo, M. | | 登録日 | 2017-08-09 | | 公開日 | 2017-12-06 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Capturing an initial intermediate during the P450nor enzymatic reaction using time-resolved XFEL crystallography and caged-substrate.
Nat Commun, 8, 2017
|
|
5Y5G
 
 | | Structure of cytochrome P450nor in NO-bound state: damaged by high-dose (5.7 MGy) X-ray | | 分子名称: | GLYCEROL, NADP nitrous oxide-forming nitric oxide reductase, NITRIC OXIDE, ... | | 著者 | Tosha, T, Nomura, T, Nishida, T, Ueno, G, Murakami, H, Yamashita, K, Hirata, K, Yamamoto, M, Ago, H, Sugimoto, H, Shiro, Y, Kubo, M. | | 登録日 | 2017-08-09 | | 公開日 | 2017-12-06 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.36 Å) | | 主引用文献 | Capturing an initial intermediate during the P450nor enzymatic reaction using time-resolved XFEL crystallography and caged-substrate.
Nat Commun, 8, 2017
|
|
