1QZX
 
 | |
3DEF
 
 | | Crystal structure of Toc33 from Arabidopsis thaliana, dimerization deficient mutant R130A | | 分子名称: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, T7I23.11 protein | | 著者 | Koenig, P, Schleiff, E, Sinning, I, Tews, I. | | 登録日 | 2008-06-10 | | 公開日 | 2008-06-24 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | On the significance of Toc-GTPase homodimers
J.Biol.Chem., 283, 2008
|
|
3DEP
 
 | | Structural basis for specific substrate recognition by the chloroplast signal recognition particle protein cpSRP43 | | 分子名称: | CHLORIDE ION, Signal recognition particle 43 kDa protein, YPGGSFDPLGLA | | 著者 | Holdermann, I, Stengel, K.F, Wild, K, Sinning, I. | | 登録日 | 2008-06-10 | | 公開日 | 2008-08-12 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structural basis for specific substrate recognition by the chloroplast signal recognition particle protein cpSRP43.
Science, 321, 2008
|
|
3DWV
 
 | |
3FEM
 
 | |
1QZW
 
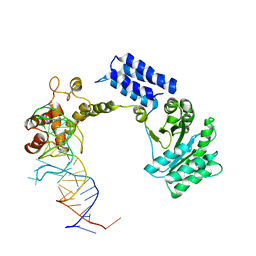 | | Crystal structure of the complete core of archaeal SRP and implications for inter-domain communication | | 分子名称: | 7S RNA, Signal recognition 54 kDa protein | | 著者 | Rosendal, K.R, Wild, K, Montoya, G, Sinning, I. | | 登録日 | 2003-09-18 | | 公開日 | 2003-11-18 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (4.1 Å) | | 主引用文献 | Crystal structure of the complete core of archaeal signal recognition particle and implications for interdomain communication
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
3DEO
 
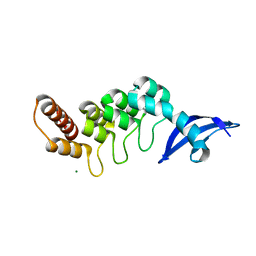 | |
4ADS
 
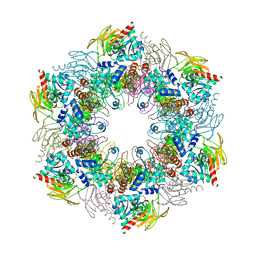 | | Crystal structure of plasmodial PLP synthase complex | | 分子名称: | PDX2 PROTEIN, PHOSPHATE ION, PYRIDOXINE BIOSYNTHETIC ENZYME PDX1 HOMOLOGUE, ... | | 著者 | Guedez, G, Sinning, I, Tews, I. | | 登録日 | 2012-01-03 | | 公開日 | 2012-01-25 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3.61 Å) | | 主引用文献 | Assembly of the Eukaryotic Plp-Synthase Complex from Plasmodium and Activation of the Pdx1 Enzyme.
Structure, 20, 2012
|
|
4ADT
 
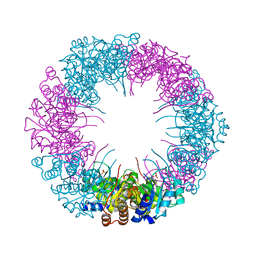 | |
4ADU
 
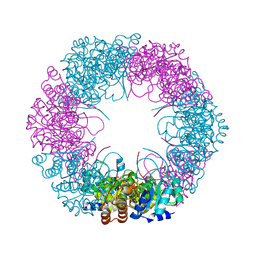 | | Crystal structure of plasmodial PLP synthase with bound R5P intermediate | | 分子名称: | 1,2-ETHANEDIOL, PYRIDOXINE BIOSYNTHETIC ENZYME PDX1 HOMOLOGUE, PUTATIVE, ... | | 著者 | Guedez, G, Sinning, I, Tews, I. | | 登録日 | 2012-01-03 | | 公開日 | 2012-01-25 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (2.44 Å) | | 主引用文献 | Assembly of the Eukaryotic Plp-Synthase Complex from Plasmodium and Activation of the Pdx1 Enzyme.
Structure, 20, 2012
|
|
2FH5
 
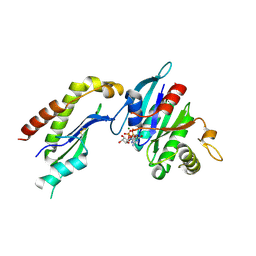 | | The Structure of the Mammalian SRP Receptor | | 分子名称: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Signal recognition particle receptor alpha subunit, ... | | 著者 | Schlenker, O, Wild, K, Sinning, I. | | 登録日 | 2005-12-23 | | 公開日 | 2006-01-31 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | The structure of the mammalian signal recognition particle (SRP) receptor as prototype for the interaction of small GTPases with Longin domains.
J.Biol.Chem., 281, 2006
|
|
2GO5
 
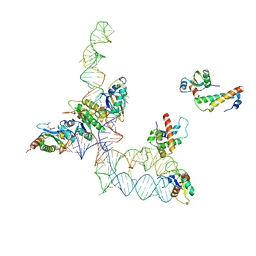 | | Structure of signal recognition particle receptor (SR) in complex with signal recognition particle (SRP) and ribosome nascent chain complex | | 分子名称: | SRP RNA, Signal recognition particle 19 kDa protein (SRP19), Signal recognition particle 54 kDa protein (SRP54), ... | | 著者 | Halic, M, Gartmann, M, Schlenker, O, Mielke, T, Pool, M.R, Sinning, I, Beckmann, R. | | 登録日 | 2006-04-12 | | 公開日 | 2006-06-13 | | 最終更新日 | 2024-10-09 | | 実験手法 | ELECTRON MICROSCOPY (7.4 Å) | | 主引用文献 | Signal Recognition Particle Receptor Exposes the Ribosomal Translocon Binding Site
Science, 312, 2006
|
|
2NV2
 
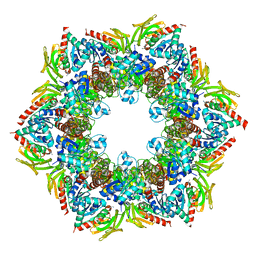 | | Structure of the PLP synthase complex Pdx1/2 (YaaD/E) from Bacillus subtilis | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, GLUTAMINE, ... | | 著者 | Strohmeier, M, Tews, I, Sinning, I. | | 登録日 | 2006-11-10 | | 公開日 | 2006-12-05 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.12 Å) | | 主引用文献 | Structure of a bacterial pyridoxal 5'-phosphate synthase complex
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2NV0
 
 | |
2NV1
 
 | | Structure of the synthase subunit Pdx1 (YaaD) of PLP synthase from Bacillus subtilis | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, MAGNESIUM ION, ... | | 著者 | Strohmeier, M, Tews, I, Sinning, I. | | 登録日 | 2006-11-10 | | 公開日 | 2006-12-05 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.08 Å) | | 主引用文献 | Structure of a bacterial pyridoxal 5'-phosphate synthase complex
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
4GMO
 
 | | Crystal structure of Syo1 | | 分子名称: | Putative uncharacterized protein | | 著者 | Bange, G, Sinning, I. | | 登録日 | 2012-08-16 | | 公開日 | 2012-10-31 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Synchronizing nuclear import of ribosomal proteins with ribosome assembly.
Science, 338, 2012
|
|
4GMQ
 
 | | Ribosome-binding domain of Zuo1 | | 分子名称: | Putative ribosome associated protein | | 著者 | Kopp, J, Bange, G, Sinning, I. | | 登録日 | 2012-08-16 | | 公開日 | 2012-12-05 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Structural characterization of a eukaryotic chaperone-the ribosome-associated complex.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4GMN
 
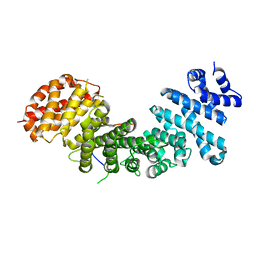 | |
4GNI
 
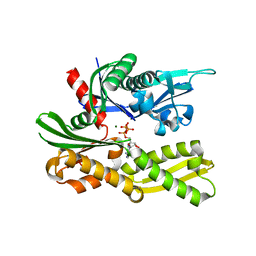 | |
3KTW
 
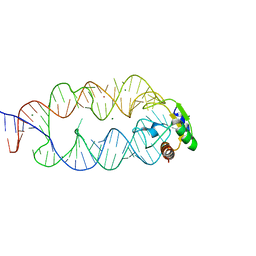 | | Crystal structure of the SRP19/S-domain SRP RNA complex of Sulfolobus solfataricus | | 分子名称: | MAGNESIUM ION, POTASSIUM ION, SRP RNA, ... | | 著者 | Wild, K, Bange, G, Bozkurt, G, Sinning, I. | | 登録日 | 2009-11-26 | | 公開日 | 2010-02-16 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Structural insights into the assembly of the human and archaeal signal recognition particles.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
5NZU
 
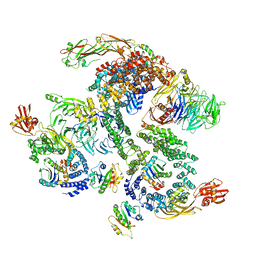 | | The structure of the COPI coat linkage II | | 分子名称: | ADP-ribosylation factor 1, Coatomer subunit alpha, Coatomer subunit beta, ... | | 著者 | Dodonova, S.O, Aderhold, P, Kopp, J, Ganeva, I, Roehling, S, Hagen, W.J.H, Sinning, I, Wieland, F, Briggs, J.A.G. | | 登録日 | 2017-05-15 | | 公開日 | 2017-06-28 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (15 Å) | | 主引用文献 | 9 angstrom structure of the COPI coat reveals that the Arf1 GTPase occupies two contrasting molecular environments.
Elife, 6, 2017
|
|
5NZV
 
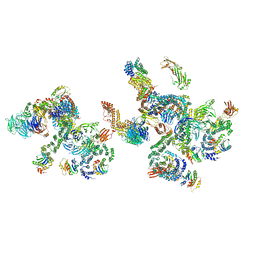 | | The structure of the COPI coat linkage IV | | 分子名称: | ADP-ribosylation factor 1, Coatomer subunit alpha, Coatomer subunit beta, ... | | 著者 | Dodonova, S.O, Aderhold, P, Kopp, J, Ganeva, I, Roehling, S, Hagen, W.J.H, Sinning, I, Wieland, F, Briggs, J.A.G. | | 登録日 | 2017-05-15 | | 公開日 | 2017-06-28 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (17.299999 Å) | | 主引用文献 | 9 angstrom structure of the COPI coat reveals that the Arf1 GTPase occupies two contrasting molecular environments.
Elife, 6, 2017
|
|
5NZS
 
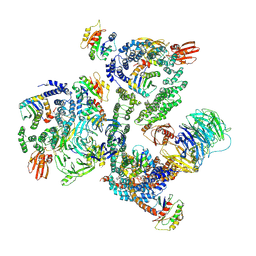 | | The structure of the COPI coat leaf in complex with the ArfGAP2 uncoating factor | | 分子名称: | ADP-ribosylation factor 1, ADP-ribosylation factor GTPase-activating protein 2, Coatomer subunit alpha, ... | | 著者 | Dodonova, S.O, Aderhold, P, Kopp, J, Ganeva, I, Roehling, S, Hagen, W.J.H, Sinning, I, Wieland, F, Briggs, J.A.G. | | 登録日 | 2017-05-15 | | 公開日 | 2017-06-28 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (10.1 Å) | | 主引用文献 | 9 angstrom structure of the COPI coat reveals that the Arf1 GTPase occupies two contrasting molecular environments.
Elife, 6, 2017
|
|
3LPZ
 
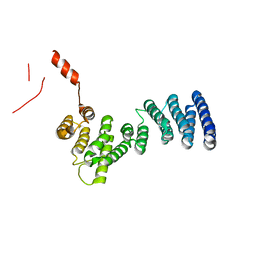 | | Crystal structure of C. therm. Get4 | | 分子名称: | Uncharacterized protein | | 著者 | Bozkurt, G, Wild, K, Sinning, I. | | 登録日 | 2010-02-08 | | 公開日 | 2010-03-16 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | The structure of Get4 reveals an alpha-solenoid fold adapted for multiple interactions in tail-anchored protein biogenesis.
Febs Lett., 584, 2010
|
|
2YHS
 
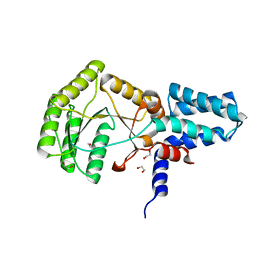 | | Structure of the E. coli SRP receptor FtsY | | 分子名称: | 1,2-ETHANEDIOL, CELL DIVISION PROTEIN FTSY | | 著者 | Stjepanovic, G, Bange, G, Wild, K, Sinning, I. | | 登録日 | 2011-05-05 | | 公開日 | 2011-05-18 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Lipids Trigger a Conformational Switch that Regulates Signal Recognition Particle (Srp)-Mediated Protein Targeting.
J.Biol.Chem., 286, 2011
|
|
