7PYZ
 
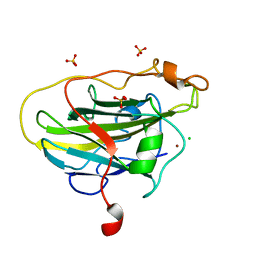 | | Structure of LPMO (expressed in E.coli) with cellotriose at 2.97x10^6 Gy | | 分子名称: | Auxiliary activity 9, CHLORIDE ION, COPPER (II) ION, ... | | 著者 | Tandrup, T, Muderspach, S.J, Banerjee, S, Ipsen, J.O, Rollan, C.H, Norholm, M.H.H, Johansen, K.S, Lo Leggio, L. | | 登録日 | 2021-10-11 | | 公開日 | 2022-08-24 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Changes in active-site geometry on X-ray photoreduction of a lytic polysaccharide monooxygenase active-site copper and saccharide binding.
Iucrj, 9, 2022
|
|
7PZ0
 
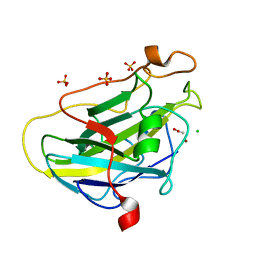 | | Structure of LPMO (expressed in E.coli) with cellotriose at 9.81x10^6 Gy | | 分子名称: | ACETATE ION, Auxiliary activity 9, CHLORIDE ION, ... | | 著者 | Tandrup, T, Muderspach, S.J, Banerjee, S, Ipsen, J.O, Rollan, C.H, Norholm, M.H.H, Johansen, K.S, Lo Leggio, L. | | 登録日 | 2021-10-11 | | 公開日 | 2022-08-24 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Changes in active-site geometry on X-ray photoreduction of a lytic polysaccharide monooxygenase active-site copper and saccharide binding.
Iucrj, 9, 2022
|
|
7PYQ
 
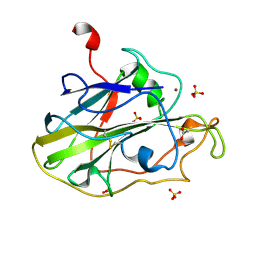 | | Structure of an LPMO (expressed in E.coli) at 6.35x10^6 Gy | | 分子名称: | ACETATE ION, Auxiliary activity 9, COPPER (II) ION, ... | | 著者 | Tandrup, T, Muderspach, S.J, Banerjee, S, Ipsen, J.O, Rollan, C.H, Norholm, M.H.H, Johansen, K.S, Lo Leggio, L. | | 登録日 | 2021-10-10 | | 公開日 | 2022-08-24 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Changes in active-site geometry on X-ray photoreduction of a lytic polysaccharide monooxygenase active-site copper and saccharide binding.
Iucrj, 9, 2022
|
|
7PXS
 
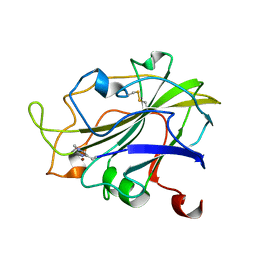 | | Room temperature X-ray structure of LPMO at 1.91x10^3 Gy | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Auxiliary activity 9, COPPER (II) ION | | 著者 | Tandrup, T, Muderspach, S.J, Banerjee, S, Lo Leggio, L. | | 登録日 | 2021-10-08 | | 公開日 | 2022-08-24 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Changes in active-site geometry on X-ray photoreduction of a lytic polysaccharide monooxygenase active-site copper and saccharide binding.
Iucrj, 9, 2022
|
|
4O6M
 
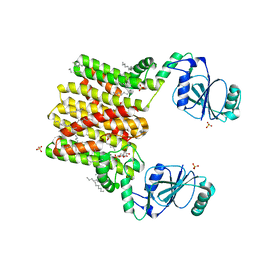 | | Structure of AF2299, a CDP-alcohol phosphotransferase (CMP-bound) | | 分子名称: | AF2299, a CDP-alcohol phosphotransferase, CALCIUM ION, ... | | 著者 | Clarke, O.B, Sciara, G, Tomasek, D, Banerjee, S, Rajashankar, K.R, Shapiro, L, Mancia, F, New York Consortium on Membrane Protein Structure (NYCOMPS) | | 登録日 | 2013-12-22 | | 公開日 | 2014-05-14 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.901 Å) | | 主引用文献 | Structural basis for catalysis in a CDP-alcohol phosphotransferase.
Nat Commun, 5, 2014
|
|
6E8C
 
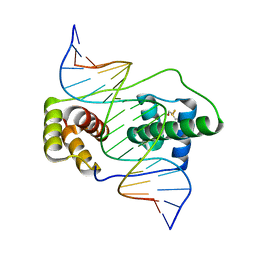 | | Crystal structure of the double homeodomain of DUX4 in complex with DNA | | 分子名称: | DNA (5'-D(*GP*CP*GP*TP*AP*AP*TP*CP*TP*AP*AP*TP*CP*AP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*TP*GP*AP*TP*TP*AP*GP*AP*TP*TP*AP*CP*GP*C)-3'), Double homeobox protein 4 | | 著者 | Lee, J.K, Bosnakovski, D, Toso, E.A, Dinh, T, Banerjee, S, Bohl, T.E, Shi, K, Kurahashi, K, Kyba, M, Aihara, H. | | 登録日 | 2018-07-27 | | 公開日 | 2018-12-26 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.12 Å) | | 主引用文献 | Crystal Structure of the Double Homeodomain of DUX4 in Complex with DNA.
Cell Rep, 25, 2018
|
|
4PWM
 
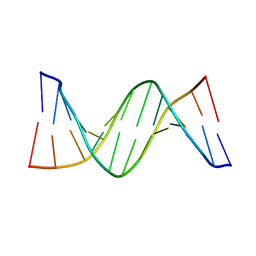 | | Crystal structure of Dickerson Drew Dodecamer with 5-carboxycytosine | | 分子名称: | 5'-[CGCGAATT(5CC)GCG]-3' | | 著者 | Szulik, M.W, Pallan, P, Banerjee, S, Voehler, M, Egli, M, Stone, M.P. | | 登録日 | 2014-03-20 | | 公開日 | 2015-02-11 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Differential stabilities and sequence-dependent base pair opening dynamics of watson-crick base pairs with 5-hydroxymethylcytosine, 5-formylcytosine, or 5-carboxylcytosine.
Biochemistry, 54, 2015
|
|
4Q7C
 
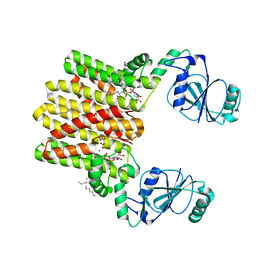 | | Structure of AF2299, a CDP-alcohol phosphotransferase | | 分子名称: | AF2299, a CDP-alcohol phosphotransferase, CALCIUM ION, ... | | 著者 | Clarke, O.B, Sciara, G, Tomasek, D, Banerjee, S, Rajashankar, K.R, Shapiro, L, Mancia, F, New York Consortium on Membrane Protein Structure (NYCOMPS) | | 登録日 | 2014-04-24 | | 公開日 | 2014-05-28 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3.102 Å) | | 主引用文献 | Structural basis for catalysis in a CDP-alcohol phosphotransferase.
Nat Commun, 5, 2014
|
|
4FM5
 
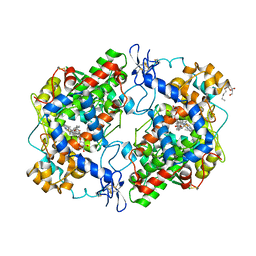 | | X-ray structure of des-methylflurbiprofen bound to murine COX-2 | | 分子名称: | (2-fluorobiphenyl-4-yl)acetic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PROTOPORPHYRIN IX CONTAINING FE, ... | | 著者 | Xu, S, Banerjee, S, Windsor, M.A, Marnett, L.J. | | 登録日 | 2012-06-15 | | 公開日 | 2012-08-29 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.81 Å) | | 主引用文献 | Substrate-Selective Inhibition of Cyclooxygenase-2: Development and Evaluation of Achiral Profen Probes.
ACS Med Chem Lett, 3, 2012
|
|
5F15
 
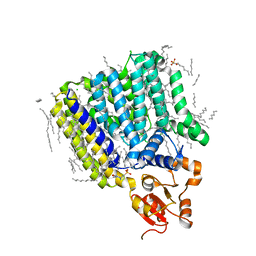 | | Crystal Structure of ArnT from Cupriavidus metallidurans bound to Undecaprenyl phosphate | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-amino-4-deoxy-L-arabinose (L-Ara4N) transferase, CHLORIDE ION, ... | | 著者 | Petrou, V.I, Clarke, O.B, Tomasek, D, Banerjee, S, Rajashankar, K.R, Mancia, F, New York Consortium on Membrane Protein Structure (NYCOMPS) | | 登録日 | 2015-11-30 | | 公開日 | 2016-02-17 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Structures of aminoarabinose transferase ArnT suggest a molecular basis for lipid A glycosylation.
Science, 351, 2016
|
|
5EZM
 
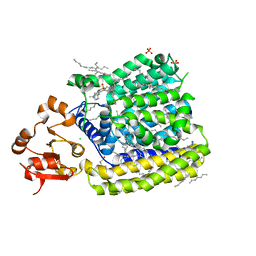 | | Crystal Structure of ArnT from Cupriavidus metallidurans in the apo state | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-amino-4-deoxy-L-arabinose transferase or related glycosyltransferases of PMT family, CHLORIDE ION, ... | | 著者 | Petrou, V.I, Clarke, O.B, Tomasek, D, Banerjee, S, Rajashankar, K.R, Mancia, F, New York Consortium on Membrane Protein Structure (NYCOMPS) | | 登録日 | 2015-11-26 | | 公開日 | 2016-02-17 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structures of aminoarabinose transferase ArnT suggest a molecular basis for lipid A glycosylation.
Science, 351, 2016
|
|
5D91
 
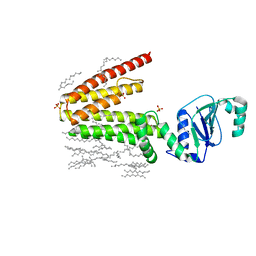 | | Structure of a phosphatidylinositolphosphate (PIP) synthase from Renibacterium Salmoninarum | | 分子名称: | AF2299 protein,Phosphatidylinositol synthase, MAGNESIUM ION, Octadecane, ... | | 著者 | Clarke, O.B, Tomasek, D.T, Jorge, C.D, Belcher Dufrisne, M, Kim, M, Banerjee, S, Rajashankar, K.R, Hendrickson, W.A, Santos, H, Mancia, F. | | 登録日 | 2015-08-18 | | 公開日 | 2015-11-11 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.501 Å) | | 主引用文献 | Structural basis for phosphatidylinositol-phosphate biosynthesis.
Nat Commun, 6, 2015
|
|
5D92
 
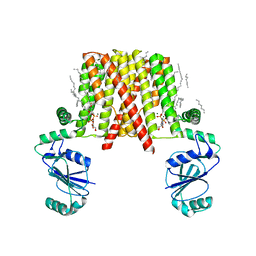 | | Structure of a phosphatidylinositolphosphate (PIP) synthase from Renibacterium Salmoninarum | | 分子名称: | 5'-O-[(R)-{[(S)-{(2R)-2,3-bis[(9E)-octadec-9-enoyloxy]propoxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]cytidine, AF2299 protein,Phosphatidylinositol synthase, MAGNESIUM ION, ... | | 著者 | Clarke, O.B, Tomasek, D.T, Jorge, C.D, Belcher Dufrisne, M, Kim, M, Banerjee, S, Rajashankar, K.R, Hendrickson, W.A, Santos, H, Mancia, F. | | 登録日 | 2015-08-18 | | 公開日 | 2015-11-04 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (3.62 Å) | | 主引用文献 | Structural basis for phosphatidylinositol-phosphate biosynthesis.
Nat Commun, 6, 2015
|
|
5HGJ
 
 | | Structure of integrin alpha1beta1 and alpha2beta1 I-domains explain differential calcium-mediated ligand recognition | | 分子名称: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | 著者 | Brown, K.L, Banerjee, S, Feigley, A, Abe, H, Blackwell, T, Zent, R, Pozzi, A, Hudson, B.H. | | 登録日 | 2016-01-08 | | 公開日 | 2017-04-12 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.399 Å) | | 主引用文献 | Salt-bridge modulates differential calcium-mediated ligand binding to integrin alpha 1- and alpha 2-I domains.
Sci Rep, 8, 2018
|
|
5HJ2
 
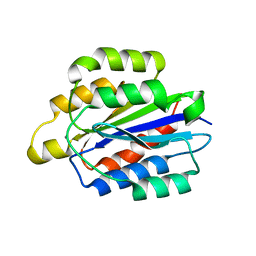 | | Integrin alpha2beta1 I-domain | | 分子名称: | CALCIUM ION, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Brown, K.L, Banerjee, S. | | 登録日 | 2016-01-12 | | 公開日 | 2017-04-12 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.153 Å) | | 主引用文献 | Salt-bridge modulates differential calcium-mediated ligand binding to integrin alpha 1- and alpha 2-I domains.
Sci Rep, 8, 2018
|
|
5ILO
 
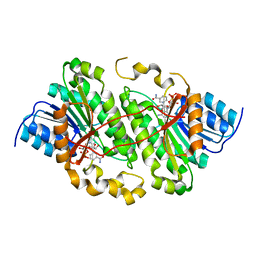 | | Crystal structure of photoreceptor dehydrogenase from Drosophila melanogaster | | 分子名称: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Photoreceptor dehydrogenase, isoform C | | 著者 | Hofmann, L, Tsybovsky, Y, Banerjee, S. | | 登録日 | 2016-03-04 | | 公開日 | 2016-11-16 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.71 Å) | | 主引用文献 | Structural Insights into the Drosophila melanogaster Retinol Dehydrogenase, a Member of the Short-Chain Dehydrogenase/Reductase Family.
Biochemistry, 55, 2016
|
|
5ILG
 
 | | Crystal structure of photoreceptor dehydrogenase from Drosophila melanogaster | | 分子名称: | 1,2-ETHANEDIOL, MAGNESIUM ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | 著者 | Hofmann, L, Tsybovsky, Y, Banerjee, S. | | 登録日 | 2016-03-04 | | 公開日 | 2016-11-16 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural Insights into the Drosophila melanogaster Retinol Dehydrogenase, a Member of the Short-Chain Dehydrogenase/Reductase Family.
Biochemistry, 55, 2016
|
|
3Q7D
 
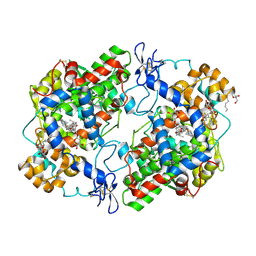 | | Structure of (R)-naproxen bound to mCOX-2. | | 分子名称: | (2R)-2-(6-methoxynaphthalen-2-yl)propanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | 著者 | Duggan, K.C, Hermanson, D.J, Musee, J, Prusakiewicz, J.J, Scheib, J, Carter, B.D, Banerjee, S, Marnett, L.J. | | 登録日 | 2011-01-04 | | 公開日 | 2011-11-09 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | (R)-Profens are substrate-selective inhibitors of endocannabinoid oxygenation by COX-2.
Nat.Chem.Biol., 7, 2011
|
|
3RR3
 
 | | Structure of (R)-flurbiprofen bound to mCOX-2 | | 分子名称: | (2R)-2-(3-fluoro-4-phenyl-phenyl)propanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, PROTOPORPHYRIN IX CONTAINING FE, ... | | 著者 | Duggan, K.C, Hermanson, D.J, Musee, J, Prusakiewicz, J.J, Scheib, J, Carter, B.D, Banerjee, S, Oates, J.A, Marnett, L.J. | | 登録日 | 2011-04-28 | | 公開日 | 2011-11-09 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.842 Å) | | 主引用文献 | (R)-Profens are substrate-selective inhibitors of endocannabinoid oxygenation by COX-2.
Nat.Chem.Biol., 7, 2011
|
|
6WQX
 
 | | Human PRPK-TPRKB complex | | 分子名称: | EKC/KEOPS complex subunit TP53RK, EKC/KEOPS complex subunit TPRKB, MAGNESIUM ION, ... | | 著者 | Li, J, Ma, X.L, Banerjee, S, Dong, Z.G. | | 登録日 | 2020-04-29 | | 公開日 | 2021-02-17 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.53 Å) | | 主引用文献 | Crystal structure of the human PRPK-TPRKB complex.
Commun Biol, 4, 2021
|
|
6X6O
 
 | | Crystal structure of T4 protein Spackle as determined by native SAD phasing | | 分子名称: | CHLORIDE ION, Protein spackle | | 著者 | Shi, K, Kurniawan, F, Banerjee, S, Moeller, N.H, Aihara, H. | | 登録日 | 2020-05-28 | | 公開日 | 2020-09-16 | | 実験手法 | X-RAY DIFFRACTION (1.52 Å) | | 主引用文献 | Crystal structure of bacteriophage T4 Spackle as determined by native SAD phasing.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
4QX6
 
 | | CRYSTAL STRUCTURE OF GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE FROM STREPTOCOCCUS AGALACTIAE NEM316 at 2.46 ANGSTROM RESOLUTION | | 分子名称: | 1,2-ETHANEDIOL, Glyceraldehyde 3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | 著者 | Ayres, C.A, Schormann, N, Banerjee, S, Chattopadhyay, D. | | 登録日 | 2014-07-18 | | 公開日 | 2014-10-15 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.46 Å) | | 主引用文献 | Structure of Streptococcus agalactiae glyceraldehyde-3-phosphate dehydrogenase holoenzyme reveals a novel surface.
Acta Crystallogr F Struct Biol Commun, 70, 2014
|
|
7K33
 
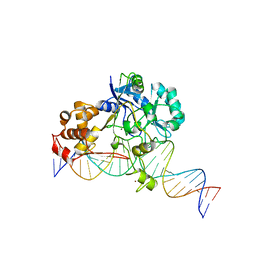 | | Crystal structure of Endonuclease Q complex with 27-mer duplex substrate with an abasic lesion at the active site | | 分子名称: | DNA (27-MER), Endonuclease Q, MAGNESIUM ION, ... | | 著者 | Shi, K, Moeller, N.M, Banerjee, S, Yin, L, Orellana, K, Aihara, H. | | 登録日 | 2020-09-10 | | 公開日 | 2021-03-17 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.11 Å) | | 主引用文献 | Structural basis for recognition of distinct deaminated DNA lesions by endonuclease Q.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7K32
 
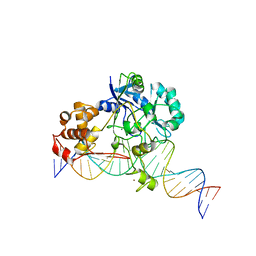 | | Crystal structure of Endonuclease Q complex with 27-mer duplex substrate with an abasic lesion at the active site | | 分子名称: | DNA (27-MER), Endonuclease Q, MAGNESIUM ION, ... | | 著者 | Shi, K, Moeller, N.M, Banerjee, S, Yin, L, Orellana, K, Aihara, H. | | 登録日 | 2020-09-10 | | 公開日 | 2021-03-17 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.11 Å) | | 主引用文献 | Structural basis for recognition of distinct deaminated DNA lesions by endonuclease Q.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7K30
 
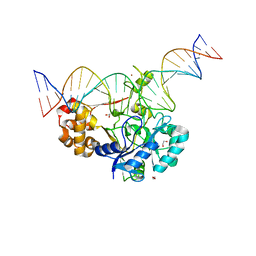 | | Crystal structure of Endonuclease Q complex with 27-mer duplex substrate with dU at the active site | | 分子名称: | 1,2-ETHANEDIOL, DNA (27-MER), Endonuclease Q, ... | | 著者 | Shi, K, Moeller, N.M, Banerjee, S, Yin, L, Orellana, K, Aihara, H. | | 登録日 | 2020-09-10 | | 公開日 | 2021-03-17 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.34 Å) | | 主引用文献 | Structural basis for recognition of distinct deaminated DNA lesions by endonuclease Q.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
