7O40
 
 | | Structural basis for VIPP1 oligomerization and maintenance of thylakoid membrane integrity | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Protein sll0617 | | 著者 | Gupta, T.K, Klumpe, S, Gries, K, Strauss, M, Rudack, T, Schuller, J.M, Schroda, M, Engel, B.D. | | 登録日 | 2021-04-03 | | 公開日 | 2021-06-30 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | Structural basis for VIPP1 oligomerization and maintenance of thylakoid membrane integrity.
Cell, 184, 2021
|
|
7O3Y
 
 | | Structural basis for VIPP1 oligomerization and maintenance of thylakoid membrane integrity | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Protein sll0617 | | 著者 | Gupta, T.K, Klumpe, S, Gries, K, Strauss, M, Rudack, T, Schuller, J.M, Schroda, M, Engel, B.D. | | 登録日 | 2021-04-03 | | 公開日 | 2021-06-30 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structural basis for VIPP1 oligomerization and maintenance of thylakoid membrane integrity.
Cell, 184, 2021
|
|
7O3Z
 
 | | Structural basis for VIPP1 oligomerization and maintenance of thylakoid membrane integrity | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Protein sll0617 | | 著者 | Gupta, T.K, Klumpe, S, Gries, K, Strauss, M, Rudack, T, Schuller, J.M, Schroda, M, Engel, B.D. | | 登録日 | 2021-04-03 | | 公開日 | 2021-06-30 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (5 Å) | | 主引用文献 | Structural basis for VIPP1 oligomerization and maintenance of thylakoid membrane integrity.
Cell, 184, 2021
|
|
7O3W
 
 | | Structural basis for VIPP1 oligomerization and maintenance of thylakoid membrane integrity | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Protein sll0617 | | 著者 | Gupta, T.K, Klumpe, S, Gries, K, Strauss, M, Rudack, T, Schuller, J.M, Schroda, M, Engel, B.D. | | 登録日 | 2021-04-03 | | 公開日 | 2021-06-30 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (4.9 Å) | | 主引用文献 | Structural basis for VIPP1 oligomerization and maintenance of thylakoid membrane integrity.
Cell, 184, 2021
|
|
7O3X
 
 | | Structural basis for VIPP1 oligomerization and maintenance of thylakoid membrane integrity | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Protein sll0617 | | 著者 | Gupta, T.K, Klumpe, S, Gries, K, Strauss, M, Rudack, T, Schuller, J.M, Schroda, M, Engel, B.D. | | 登録日 | 2021-04-03 | | 公開日 | 2021-06-30 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Structural basis for VIPP1 oligomerization and maintenance of thylakoid membrane integrity.
Cell, 184, 2021
|
|
5JRS
 
 | |
7JTM
 
 | | CRYSTAL STRUCTURE OF RAR-RELATED ORPHAN RECEPTOR C (NHIS-RORGT(244-487)-L6-SRC1(6T78-692) IN COMPLEX WITH A TRICYCLIC SULFONE RORGT INVERSE AGONIST | | 分子名称: | Nuclear receptor ROR-gamma, trans-4-[(3aR,9bR)-8-cyano-9b-[(4-fluorophenyl)sulfonyl]-7-(1,1,1,2,3,3,3-heptafluoropropan-2-yl)-1,2,3a,4,5,9b-hexahydro-3H-benzo[e]indole-3-carbonyl]cyclohexane-1-carboxylic acid | | 著者 | Sack, J.S. | | 登録日 | 2020-08-18 | | 公開日 | 2020-09-16 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.43 Å) | | 主引用文献 | Tricyclic sulfones as potent, selective and efficacious ROR gamma t inverse agonists - Exploring C6 and C8 SAR using late-stage functionalization.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
7A1G
 
 | | Structure of a crosslinked yeast ABCE1-bound 43S pre-initiation complex | | 分子名称: | 18S ribosomal RNA, 40S ribosomal protein S0-A, 40S ribosomal protein S1-A, ... | | 著者 | Mackens-Kiani, T, Kratzat, H, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | 登録日 | 2020-08-13 | | 公開日 | 2020-10-14 | | 最終更新日 | 2023-11-29 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | A structural inventory of native ribosomal ABCE1-43S pre-initiation complexes.
Embo J., 40, 2021
|
|
2OFZ
 
 | | Ultrahigh Resolution Crystal Structure of RNA Binding Domain of SARS Nucleopcapsid (N Protein) at 1.1 Angstrom Resolution in Monoclinic Form. | | 分子名称: | 1,2-ETHANEDIOL, Nucleocapsid protein | | 著者 | Saikatendu, K, Joseph, J, Subramanian, V, Neuman, B, Buchmeier, M, Stevens, R.C, Kuhn, P. | | 登録日 | 2007-01-04 | | 公開日 | 2007-04-03 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.17 Å) | | 主引用文献 | Ribonucleocapsid formation of severe acute respiratory syndrome coronavirus through molecular action of the N-terminal domain of N protein.
J.Virol., 81, 2007
|
|
2OG3
 
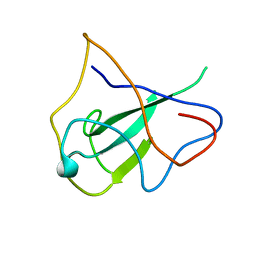 | | structure of the rna binding domain of n protein from SARS coronavirus in cubic crystal form | | 分子名称: | Nucleocapsid protein | | 著者 | Saikatendu, K, Joseph, J, Subramanian, V, Neuman, B, Buchmeier, M, Stevens, R.C, Kuhn, P. | | 登録日 | 2007-01-05 | | 公開日 | 2007-04-03 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Ribonucleocapsid formation of severe acute respiratory syndrome coronavirus through molecular action of the N-terminal domain of N protein.
J.Virol., 81, 2007
|
|
2OZK
 
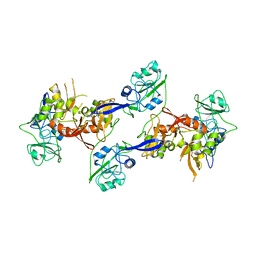 | | Structure of an N-Terminal Truncated Form of Nendou (NSP15) From SARS-CORONAVIRUS | | 分子名称: | Uridylate-specific endoribonuclease | | 著者 | Saikatendu, K, Joseph, J, Subramanian, V, Neuman, B, Buchmeier, M, Stevens, R.C, Kuhn, P. | | 登録日 | 2007-02-26 | | 公開日 | 2007-05-29 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Crystal structure of a monomeric form of severe acute respiratory syndrome coronavirus endonuclease nsp15 suggests a role for hexamerization as an allosteric switch.
J.Virol., 81, 2007
|
|
6ZQE
 
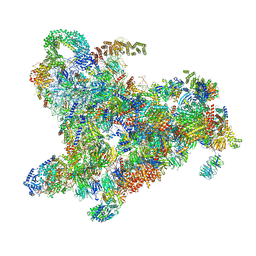 | | Cryo-EM structure of the 90S pre-ribosome from Saccharomyces cerevisiae, state Dis-A (Poly-Ala) | | 分子名称: | 13 kDa ribonucleoprotein-associated protein, 18S rRNA, 40S ribosomal protein S1-A, ... | | 著者 | Cheng, J, Lau, B, Venuta, G.L, Berninghausen, O, Hurt, E, Beckmann, R. | | 登録日 | 2020-07-09 | | 公開日 | 2020-09-23 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (7.1 Å) | | 主引用文献 | 90 S pre-ribosome transformation into the primordial 40 S subunit.
Science, 369, 2020
|
|
6ZQC
 
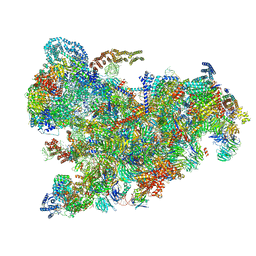 | | Cryo-EM structure of the 90S pre-ribosome from Saccharomyces cerevisiae, state Pre-A1 | | 分子名称: | 13 kDa ribonucleoprotein-associated protein, 18S rRNA, 40S ribosomal protein S1-A, ... | | 著者 | Cheng, J, Lau, B, Venuta, G.L, Berninghausen, O, Hurt, E, Beckmann, R. | | 登録日 | 2020-07-09 | | 公開日 | 2020-09-23 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | 90 S pre-ribosome transformation into the primordial 40 S subunit.
Science, 369, 2020
|
|
6ZQG
 
 | | Cryo-EM structure of the 90S pre-ribosome from Saccharomyces cerevisiae, state Dis-C | | 分子名称: | 18S rRNA, 40S ribosomal protein S1-A, 40S ribosomal protein S11-A, ... | | 著者 | Cheng, J, Lau, B, Venuta, G.L, Berninghausen, O, Hurt, E, Beckmann, R. | | 登録日 | 2020-07-09 | | 公開日 | 2020-09-23 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | 90 S pre-ribosome transformation into the primordial 40 S subunit.
Science, 369, 2020
|
|
6ZQB
 
 | | Cryo-EM structure of the 90S pre-ribosome from Saccharomyces cerevisiae, state B2 | | 分子名称: | 13 kDa ribonucleoprotein-associated protein, 18S rRNA, 40S ribosomal protein S1-A, ... | | 著者 | Cheng, J, Lau, B, Venuta, G.L, Berninghausen, O, Hurt, E, Beckmann, R. | | 登録日 | 2020-07-09 | | 公開日 | 2020-09-23 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | 90 S pre-ribosome transformation into the primordial 40 S subunit.
Science, 369, 2020
|
|
6ZQF
 
 | | Cryo-EM structure of the 90S pre-ribosome from Saccharomyces cerevisiae, state Dis-B (Poly-Ala) | | 分子名称: | 18S rRNA, 40S ribosomal protein S1-A, 40S ribosomal protein S11-A, ... | | 著者 | Cheng, J, Lau, B, Venuta, G.L, Berninghausen, O, Hurt, E, Beckmann, R. | | 登録日 | 2020-07-09 | | 公開日 | 2020-09-23 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (4.9 Å) | | 主引用文献 | 90 S pre-ribosome transformation into the primordial 40 S subunit.
Science, 369, 2020
|
|
3HPI
 
 | |
6ZQD
 
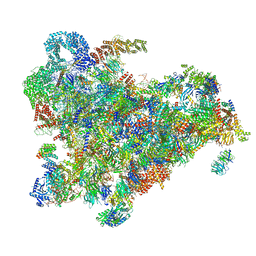 | | Cryo-EM structure of the 90S pre-ribosome from Saccharomyces cerevisiae, state Post-A1 | | 分子名称: | 13 kDa ribonucleoprotein-associated protein, 18S rRNA, 40S ribosomal protein S1-A, ... | | 著者 | Cheng, J, Lau, B, Venuta, G.L, Berninghausen, O, Hurt, E, Beckmann, R. | | 登録日 | 2020-07-09 | | 公開日 | 2020-09-23 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | 90 S pre-ribosome transformation into the primordial 40 S subunit.
Science, 369, 2020
|
|
4DXB
 
 | |
4DXC
 
 | |
