5FJ0
 
 | |
5FK6
 
 | |
5FK1
 
 | |
5FK3
 
 | |
5FK4
 
 | |
5FJC
 
 | |
5FKG
 
 | |
5FKE
 
 | |
5FK2
 
 | |
5FKF
 
 | |
5FKD
 
 | |
5FJ1
 
 | |
5FK5
 
 | |
5FKH
 
 | |
5FJ4
 
 | |
5G4U
 
 | |
5G4T
 
 | |
5G4V
 
 | |
6HCT
 
 | |
6HC5
 
 | |
7EAG
 
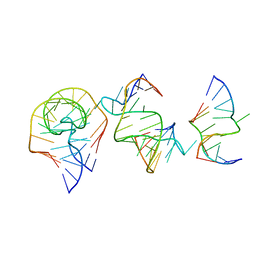 | | Crystal structure of the RAGATH-18 k-turn | | 分子名称: | RNA (5'-R(*GP*UP*CP*UP*AP*UP*GP*AP*AP*GP*GP*CP*UP*GP*GP*AP*GP*AP*C)-3') | | 著者 | Huang, L, Lilley, D.M.J. | | 登録日 | 2021-03-07 | | 公開日 | 2021-06-02 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structure and folding of four putative kink turns identified in structured RNA species in a test of structural prediction rules.
Nucleic Acids Res., 49, 2021
|
|
7EAF
 
 | |
1DUS
 
 | | MJ0882-A hypothetical protein from M. jannaschii | | 分子名称: | MJ0882 | | 著者 | Hung, L, Huang, L, Kim, R, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | 登録日 | 2000-01-18 | | 公開日 | 2000-07-19 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structure-based experimental confirmation of biochemical function to a methyltransferase, MJ0882, from hyperthermophile Methanococcus jannaschii
J.STRUCT.FUNCT.GENOM., 2, 2002
|
|
6Q8U
 
 | |
6Q8V
 
 | |
