6INF
 
 | | a glycosyltransferase complex with UDP | | 分子名称: | UDP-glycosyltransferase 76G1, URIDINE-5'-DIPHOSPHATE | | 著者 | Zhu, X, Yang, T, Naismith, J.H. | | 登録日 | 2018-10-25 | | 公開日 | 2019-07-31 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.69 Å) | | 主引用文献 | Hydrophobic recognition allows the glycosyltransferase UGT76G1 to catalyze its substrate in two orientations.
Nat Commun, 10, 2019
|
|
7E9S
 
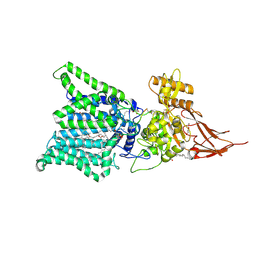 | | Archaeal oligosaccharyltransferase AglB from Archaeoglobus fulgidus in complex with an inhibitory peptide and a dolichol-phosphate | | 分子名称: | (2R)-2,3-DIHYDROXYPROPYL (7Z)-TETRADEC-7-ENOATE, DI(HYDROXYETHYL)ETHER, Dolichyl-phosphooligosaccharide-protein glycotransferase 3, ... | | 著者 | Taguchi, Y, Hirata, K, Kohda, D. | | 登録日 | 2021-03-05 | | 公開日 | 2021-09-08 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | The structure of an archaeal oligosaccharyltransferase provides insight into the strict exclusion of proline from the N-glycosylation sequon.
Commun Biol, 4, 2021
|
|
6INI
 
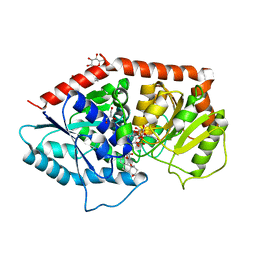 | | a glycosyltransferase complex with UDP and the product | | 分子名称: | (8alpha,9beta,10alpha,13alpha)-13-{[beta-D-glucopyranosyl-(1->2)-[beta-D-glucopyranosyl-(1->3)]-beta-D-glucopyranosyl]oxy}kaur-16-en-18-oic acid, 1-O-[(8alpha,9beta,10alpha,13alpha)-13-(beta-D-glucopyranosyloxy)-18-oxokaur-16-en-18-yl]-beta-D-glucopyranose, GLYCEROL, ... | | 著者 | Zhu, X, Yang, T, Naismith, J.H. | | 登録日 | 2018-10-25 | | 公開日 | 2019-07-31 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Hydrophobic recognition allows the glycosyltransferase UGT76G1 to catalyze its substrate in two orientations.
Nat Commun, 10, 2019
|
|
4L3P
 
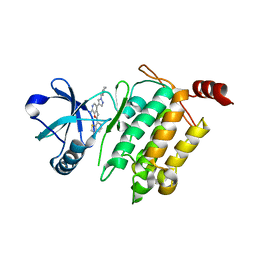 | | Crystal Structure of 2-(1-benzothiophen-7-yl)-4-[1-(piperidin-4-yl)-1H-pyrazol-4-yl]furo[2,3-c]pyridin-7-amine bound to TAK1-TAB1 | | 分子名称: | 2-(1-benzothiophen-7-yl)-4-[1-(piperidin-4-yl)-1H-pyrazol-4-yl]furo[2,3-c]pyridin-7-amine, Mitogen-activated protein kinase kinase kinase 7, TGF-beta-activated kinase 1 and MAP3K7-binding protein 1 chimera | | 著者 | Wang, J, Hornberger, K.R, Crew, A.P, Steinbacher, S, Maskos, K, Moertl, M. | | 登録日 | 2013-06-06 | | 公開日 | 2013-06-19 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.68 Å) | | 主引用文献 | Discovery and optimization of 7-aminofuro[2,3-c]pyridine inhibitors of TAK1.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
2RLL
 
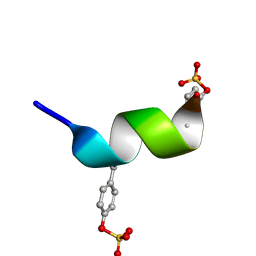 | | CCR5 Nt(7-15) | | 分子名称: | 9-mer from C-C chemokine receptor type 5 | | 著者 | Bewley, C.A, Lam, S.N. | | 登録日 | 2007-07-21 | | 公開日 | 2007-09-25 | | 最終更新日 | 2023-11-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structures of the CCR5 N terminus and of a tyrosine-sulfated antibody with HIV-1 gp120 and CD4
Science, 317, 2007
|
|
2ZXC
 
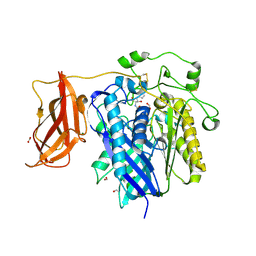 | | Ceramidase complexed with C2 | | 分子名称: | DIMETHYL SULFOXIDE, FORMIC ACID, MAGNESIUM ION, ... | | 著者 | Okano, H, Inoue, T, Okino, N, Kakuta, Y, Matsumura, H, Ito, M. | | 登録日 | 2008-12-22 | | 公開日 | 2009-02-03 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Mechanistic insights into the hydrolysis and synthesis of ceramide by neutral ceramidase.
J.Biol.Chem., 284, 2009
|
|
5GZ6
 
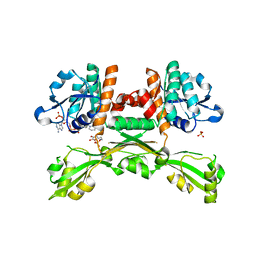 | | Structure of D-amino acid dehydrogenase in complex with NADPH and 2-keto-6-aminocapronic acid | | 分子名称: | 6-azanyl-2-oxidanylidene-hexanoic acid, ACETATE ION, Meso-diaminopimelate D-dehydrogenase, ... | | 著者 | Sakuraba, H, Seto, T, Hayashi, J, Akita, H, Yoneda, K, Ohshima, T. | | 登録日 | 2016-09-26 | | 公開日 | 2017-04-12 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.74 Å) | | 主引用文献 | Structure-Based Engineering of an Artificially Generated NADP+-Dependent d-Amino Acid Dehydrogenase
Appl. Environ. Microbiol., 83, 2017
|
|
5GZ3
 
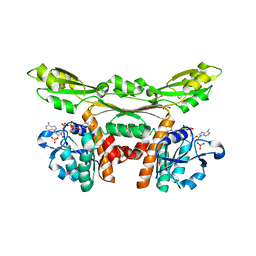 | | Structure of D-amino acid dehydrogenase in complex with NADP | | 分子名称: | 1,2-ETHANEDIOL, Meso-diaminopimelate D-dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Sakuraba, H, Seto, T, Hayashi, J, Akita, H, Yoneda, K, Ohshima, T. | | 登録日 | 2016-09-26 | | 公開日 | 2017-04-12 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.59 Å) | | 主引用文献 | Structure-Based Engineering of an Artificially Generated NADP+-Dependent d-Amino Acid Dehydrogenase
Appl. Environ. Microbiol., 83, 2017
|
|
5FLY
 
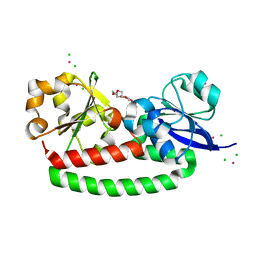 | | The FhuD protein from S.pseudintermedius | | 分子名称: | 2,5,8,11,14,17,20,23-OCTAOXAPENTACOSAN-25-OL, CADMIUM ION, CHLORIDE ION, ... | | 著者 | Malito, E. | | 登録日 | 2015-10-29 | | 公開日 | 2015-12-02 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.598 Å) | | 主引用文献 | Crystal Structure of Fhud at 1.6 Angstrom Resolution: A Ferrichrome-Binding Protein from the Animal and Human Pathogen Staphylococcus Pseudintermedius
Acta Crystallogr.,Sect.F, 72, 2016
|
|
2Z98
 
 | |
2Z9C
 
 | |
2Z9D
 
 | |
5GZ1
 
 | | Structure of substrate/cofactor-free D-amino acid dehydrogenase | | 分子名称: | Meso-diaminopimelate D-dehydrogenase | | 著者 | Sakuraba, H, Seto, T, Hayashi, J, Akita, H, Yoneda, K, Ohshima, T. | | 登録日 | 2016-09-26 | | 公開日 | 2017-04-12 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | Structure-Based Engineering of an Artificially Generated NADP+-Dependent d-Amino Acid Dehydrogenase
Appl. Environ. Microbiol., 83, 2017
|
|
4L52
 
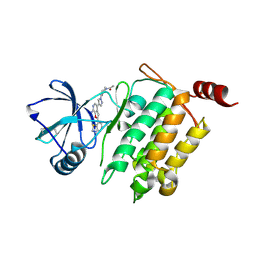 | | Crystal Structure of 1-(4-{4-[7-amino-2-(1,2,3-benzothiadiazol-7-yl)furo[2,3-c]pyridin-4-yl]-1H-pyrazol-1-yl}piperidin-1-yl)ethan-1-one bound to TAK1-TAB1 | | 分子名称: | 1-(4-{4-[7-amino-2-(1,2,3-benzothiadiazol-7-yl)furo[2,3-c]pyridin-4-yl]-1H-pyrazol-1-yl}piperidin-1-yl)ethanone, Mitogen-activated protein kinase kinase kinase 7, TGF-beta-activated kinase 1 and MAP3K7-binding protein 1 chimera | | 著者 | Wang, J, Hornberger, K.R, Crew, A.P, Jestel, A, Maskos, K, Moertl, M. | | 登録日 | 2013-06-10 | | 公開日 | 2013-07-03 | | 最終更新日 | 2017-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.54 Å) | | 主引用文献 | Discovery and optimization of 7-aminofuro[2,3-c]pyridine inhibitors of TAK1.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
8IJN
 
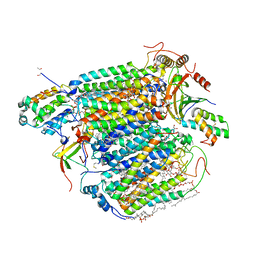 | | Bovine Heart Cytochrome c Oxidase in the Nitric Oxide-Bound Fully Reduced State at 100 K | | 分子名称: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | 著者 | Tsukihara, T, Shimada, A, Muramoto, K. | | 登録日 | 2023-02-27 | | 公開日 | 2023-04-12 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Bovine cytochrome c oxidase structures enable O2 reduction with minimization of reactive oxygens and provide a proton-pumping gate.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
5ZFU
 
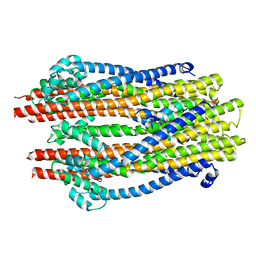 | | Structure of the ExbB/ExbD hexameric complex (ExbB6ExbD3TM) | | 分子名称: | 22-mer peptide from Biopolymer transport protein ExbD, Biopolymer transport protein ExbB | | 著者 | Yonekura, K, Yamashita, Y, Matsuoka, R, Maki-Yonekura, S. | | 登録日 | 2018-03-07 | | 公開日 | 2018-05-09 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (6.7 Å) | | 主引用文献 | Hexameric and pentameric complexes of the ExbBD energizer in the Ton system.
Elife, 7, 2018
|
|
5ZFP
 
 | |
5ZFV
 
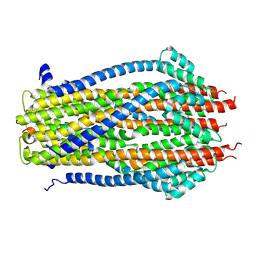 | | Structure of the ExbB/ExbD pentameric complex (ExbB5ExbD1TM) | | 分子名称: | 22-mer peptide from Biopolymer transport protein ExbD, Biopolymer transport protein ExbB | | 著者 | Yonekura, K, Yamashita, Y, Matsuoka, R, Maki-Yonekura, S. | | 登録日 | 2018-03-07 | | 公開日 | 2018-05-09 | | 実験手法 | ELECTRON MICROSCOPY (7.1 Å) | | 主引用文献 | Hexameric and pentameric complexes of the ExbBD energizer in the Ton system.
Elife, 7, 2018
|
|
6ING
 
 | |
2Z9B
 
 | |
4EJN
 
 | | Crystal structure of autoinhibited form of AKT1 in complex with N-(4-(5-(3-acetamidophenyl)-2-(2-aminopyridin-3-yl)-3H-imidazo[4,5-b]pyridin-3-yl)benzyl)-3-fluorobenzamide | | 分子名称: | 1,2-ETHANEDIOL, 2-BUTANOL, N-(4-{5-[3-(acetylamino)phenyl]-2-(2-aminopyridin-3-yl)-3H-imidazo[4,5-b]pyridin-3-yl}benzyl)-3-fluorobenzamide, ... | | 著者 | Eathiraj, S. | | 登録日 | 2012-04-06 | | 公開日 | 2012-05-23 | | 最終更新日 | 2013-01-02 | | 実験手法 | X-RAY DIFFRACTION (2.19 Å) | | 主引用文献 | Discovery and optimization of a series of 3-(3-phenyl-3H-imidazo[4,5-b]pyridin-2-yl)pyridin-2-amines: orally bioavailable, selective, and potent ATP-independent Akt inhibitors.
J.Med.Chem., 55, 2012
|
|
2ZWS
 
 | | Crystal Structure Analysis of neutral ceramidase from Pseudomonas aeruginosa | | 分子名称: | FORMIC ACID, GLYCEROL, MAGNESIUM ION, ... | | 著者 | Kakuta, Y, Okino, N, Inoue, T, Okano, H, Ito, M. | | 登録日 | 2008-12-17 | | 公開日 | 2009-03-03 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Mechanistic insights into the hydrolysis and synthesis of ceramide by neutral ceramidase.
J.Biol.Chem., 284, 2009
|
|
4G3G
 
 | |
4OBU
 
 | |
4G3E
 
 | |
