5Z1N
 
 | | Crystal structure of C terminal region of G-protein interacting protein 1 (Gip1) from Dictyostelium discoideum | | 分子名称: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, DI-PALMITOYL-3-SN-PHOSPHATIDYLETHANOLAMINE, G-protein interacting protein 1, ... | | 著者 | Miyagawa, T, Koteishi, H, Kamimura, Y, Miyanaga, Y, Takeshita, K, Nakagawa, A, Ueda, M. | | 登録日 | 2017-12-27 | | 公開日 | 2018-10-17 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.949 Å) | | 主引用文献 | Structural basis of Gip1 for cytosolic sequestration of G protein in wide-range chemotaxis
Nat Commun, 9, 2018
|
|
5Z39
 
 | | Crystal structure of C terminal region of G-protein interacting protein 1 (Gip1) from Dictyostelium discoideum form II | | 分子名称: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, DI-PALMITOYL-3-SN-PHOSPHATIDYLETHANOLAMINE, G-protein interacting protein 1, ... | | 著者 | Miyagawa, T, Koteishi, H, Kamimura, Y, Miyanaga, Y, Takeshita, K, Nakagawa, A, Ueda, M. | | 登録日 | 2018-01-05 | | 公開日 | 2018-10-17 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.74 Å) | | 主引用文献 | Structural basis of Gip1 for cytosolic sequestration of G protein in wide-range chemotaxis
Nat Commun, 9, 2018
|
|
3VUS
 
 | | Escherichia coli PgaB N-terminal domain | | 分子名称: | ACETATE ION, MERCURY (II) ION, Poly-beta-1,6-N-acetyl-D-glucosamine N-deacetylase, ... | | 著者 | Nishiyama, T, Noguchi, H, Yoshida, H, Park, S.-Y, Tame, J.R.H. | | 登録日 | 2012-07-05 | | 公開日 | 2012-11-14 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | The structure of the deacetylase domain of Escherichia coli PgaB, an enzyme required for biofilm formation: a circularly permuted member of the carbohydrate esterase 4 family
Acta Crystallogr.,Sect.D, 69, 2013
|
|
8GS2
 
 | | Structure of the Cas7-11-Csx29-guide RNA-target RNA (non-matching PFS) complex | | 分子名称: | ADENOSINE MONOPHOSPHATE, CHAT domain-containing protein, CRISPR-associated RAMP family protein, ... | | 著者 | Kato, K, Okazaki, S, Ishikawa, J, Isayama, Y, Nishizawa, T, Nishimasu, H. | | 登録日 | 2022-09-04 | | 公開日 | 2022-11-09 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (2.84 Å) | | 主引用文献 | RNA-triggered protein cleavage and cell growth arrest by the type III-E CRISPR nuclease-protease.
Science, 378, 2022
|
|
3WHJ
 
 | | Crystal structure of Nas2 N-terminal domain | | 分子名称: | CADMIUM ION, Probable 26S proteasome regulatory subunit p27, SULFATE ION | | 著者 | Satoh, T, Saeki, Y, Hiromoto, T, Wang, Y.-H, Uekusa, Y, Yagi, H, Yoshihara, H, Yagi-Utsumi, M, Mizushima, T, Tanaka, K, Kato, K. | | 登録日 | 2013-08-26 | | 公開日 | 2014-03-26 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Structural basis for proteasome formation controlled by an assembly chaperone nas2.
Structure, 22, 2014
|
|
3WHK
 
 | | Crystal structure of PAN-Rpt5C chimera | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Proteasome-activating nucleotidase, 26S protease regulatory subunit 6A | | 著者 | Satoh, T, Saeki, Y, Hiromoto, T, Wang, Y.-H, Uekusa, Y, Yagi, H, Yoshihara, H, Yagi-Utsumi, M, Mizushima, T, Tanaka, K, Kato, K. | | 登録日 | 2013-08-26 | | 公開日 | 2014-03-26 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structural basis for proteasome formation controlled by an assembly chaperone nas2.
Structure, 22, 2014
|
|
3WXA
 
 | | X-ray crystal structural analysis of the complex between ALG-2 and Sec31A peptide | | 分子名称: | Programmed cell death protein 6, Protein transport protein Sec31A, ZINC ION | | 著者 | Takahashi, T, Suzuki, H, Kawasaki, M, Shibata, H, Wakatsuki, S, Maki, M. | | 登録日 | 2014-07-29 | | 公開日 | 2015-03-11 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.36 Å) | | 主引用文献 | Structural Analysis of the Complex between Penta-EF-Hand ALG-2 Protein and Sec31A Peptide Reveals a Novel Target Recognition Mechanism of ALG-2
Int J Mol Sci, 16, 2015
|
|
3WHL
 
 | | Crystal structure of Nas2 N-terminal domain complexed with PAN-Rpt5C chimera | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Probable 26S proteasome regulatory subunit p27, Proteasome-activating nucleotidase, ... | | 著者 | Satoh, T, Saeki, Y, Hiromoto, T, Wang, Y.-H, Uekusa, Y, Yagi, H, Yoshihara, H, Yagi-Utsumi, M, Mizushima, T, Tanaka, K, Kato, K. | | 登録日 | 2013-08-26 | | 公開日 | 2014-03-26 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (4 Å) | | 主引用文献 | Structural basis for proteasome formation controlled by an assembly chaperone nas2.
Structure, 22, 2014
|
|
3X3Y
 
 | | Copper amine oxidase from Arthrobacter globiformis anaerobically reduced by histamine | | 分子名称: | COPPER (II) ION, GLYCEROL, POTASSIUM ION, ... | | 著者 | Okajima, T, Nakanishi, S, Murakawa, T, Kataoka, M, Hayashi, H, Hamaguchi, A, Nakai, T, Kawano, Y, Yamaguchi, H, Tanizawa, K. | | 登録日 | 2015-03-10 | | 公開日 | 2015-08-19 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.499 Å) | | 主引用文献 | Probing the Catalytic Mechanism of Copper Amine Oxidase from Arthrobacter globiformis with Halide Ions.
J.Biol.Chem., 290, 2015
|
|
3X42
 
 | | Crystal structure of copper amine oxidase from Arthrobacter globiformis in the presence of sodium bromide | | 分子名称: | BROMIDE ION, COPPER (II) ION, GLYCEROL, ... | | 著者 | Okajima, T, Nakanishi, S, Murakawa, T, Kataoka, M, Hayashi, H, Hamaguchi, A, Nakai, T, Kawano, Y, Yamaguchi, H, Tanizawa, K. | | 登録日 | 2015-03-10 | | 公開日 | 2015-08-19 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.875 Å) | | 主引用文献 | Probing the Catalytic Mechanism of Copper Amine Oxidase from Arthrobacter globiformis with Halide Ions.
J.Biol.Chem., 290, 2015
|
|
3X41
 
 | | Copper amine oxidase from Arthrobacter globiformis: Product Schiff-base form produced by anaerobic reduction in the presence of sodium bromide | | 分子名称: | BROMIDE ION, COPPER (II) ION, GLYCEROL, ... | | 著者 | Okajima, T, Nakanishi, S, Murakawa, T, Kataoka, M, Hayashi, H, Hamaguchi, A, Nakai, T, Kawano, Y, Yamaguchi, H, Tanizawa, K. | | 登録日 | 2015-03-10 | | 公開日 | 2015-08-19 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.87 Å) | | 主引用文献 | Probing the Catalytic Mechanism of Copper Amine Oxidase from Arthrobacter globiformis with Halide Ions.
J.Biol.Chem., 290, 2015
|
|
3X3X
 
 | | Copper amine oxidase from Arthrobacter globiformis anaerobically reduced by phenylethylamine | | 分子名称: | 2-PHENYL-ETHANOL, COPPER (II) ION, GLYCEROL, ... | | 著者 | Okajima, T, Nakanishi, S, Murakawa, T, Kataoka, M, Hayashi, H, Hamaguchi, A, Nakai, T, Kawano, Y, Yamaguchi, H, Tanizawa, K. | | 登録日 | 2015-03-10 | | 公開日 | 2015-08-19 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.57 Å) | | 主引用文献 | Probing the Catalytic Mechanism of Copper Amine Oxidase from Arthrobacter globiformis with Halide Ions.
J.Biol.Chem., 290, 2015
|
|
3X3Z
 
 | | Copper amine oxidase from Arthrobacter globiformis: Aminoresorcinol form produced by anaerobic reduction with ethylamine hydrochloride | | 分子名称: | CHLORIDE ION, COPPER (II) ION, GLYCEROL, ... | | 著者 | Okajima, T, Nakanishi, S, Murakawa, T, Kataoka, M, Hayashi, H, Hamaguchi, A, Nakai, T, Kawano, Y, Yamaguchi, H, Tanizawa, K. | | 登録日 | 2015-03-10 | | 公開日 | 2015-08-19 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.51 Å) | | 主引用文献 | Probing the Catalytic Mechanism of Copper Amine Oxidase from Arthrobacter globiformis with Halide Ions.
J.Biol.Chem., 290, 2015
|
|
3X40
 
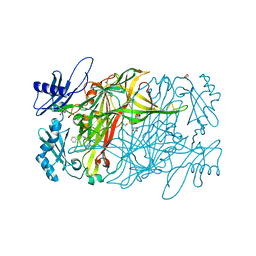 | | Copper amine oxidase from Arthrobacter globiformis: Product Schiff-base form produced by anaerobic reduction in the presence of sodium chloride | | 分子名称: | CHLORIDE ION, COPPER (II) ION, GLYCEROL, ... | | 著者 | Okajima, T, Nakanishi, S, Murakawa, T, Kataoka, M, Hayashi, H, Hamaguchi, A, Nakai, T, Kawano, Y, Yamaguchi, H, Tanizawa, K. | | 登録日 | 2015-03-10 | | 公開日 | 2015-08-19 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Probing the Catalytic Mechanism of Copper Amine Oxidase from Arthrobacter globiformis with Halide Ions.
J.Biol.Chem., 290, 2015
|
|
6MCR
 
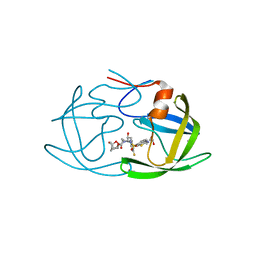 | | X-ray crystal structure of wild type HIV-1 protease in complex with GRL-001 | | 分子名称: | (3S,3aR,5R,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl [(2S,3R)-4-[{[2-(cyclopropylamino)-1,3-benzothiazol-6-yl]sulfonyl}(2-methylpropyl)amino]-1-(3-fluorophenyl)-3-hydroxybutan-2-yl]carbamate, 1,2-ETHANEDIOL, Protease | | 著者 | Bulut, H, Hayashi, H, Hattori, S.I, Aoki, M, Das, D, Ghosh, A.K, Mitsuya, H. | | 登録日 | 2018-09-02 | | 公開日 | 2019-04-24 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.48 Å) | | 主引用文献 | Halogen Bond Interactions of Novel HIV-1 Protease Inhibitors (PI) (GRL-001-15 and GRL-003-15) with the Flap of Protease Are Critical for Their Potent Activity against Wild-Type HIV-1 and Multi-PI-Resistant Variants.
Antimicrob.Agents Chemother., 63, 2019
|
|
6MCS
 
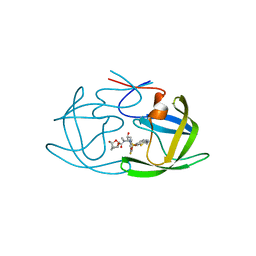 | | X-ray crystal structure of wild type HIV-1 protease in complex with GRL-003 | | 分子名称: | (3S,3aR,5R,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl [(2S,3R)-4-[{[2-(cyclopropylamino)-1,3-benzothiazol-6-yl]sulfonyl}(2-methylpropyl)amino]-1-(4-fluorophenyl)-3-hydroxybutan-2-yl]carbamate, Protease | | 著者 | Bulut, H, Hayashi, H, Hattori, S.I, Aoki, M, Das, D, Ghosh, A.K, Mitsuya, H. | | 登録日 | 2018-09-02 | | 公開日 | 2019-04-24 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.52 Å) | | 主引用文献 | Halogen Bond Interactions of Novel HIV-1 Protease Inhibitors (PI) (GRL-001-15 and GRL-003-15) with the Flap of Protease Are Critical for Their Potent Activity against Wild-Type HIV-1 and Multi-PI-Resistant Variants.
Antimicrob.Agents Chemother., 63, 2019
|
|
6LMT
 
 | | Cryo-EM structure of the killifish CALHM1 | | 分子名称: | CHOLESTEROL HEMISUCCINATE, Calcium homeostasis modulator 1 | | 著者 | Demura, K, Kusakizako, T, Shihoya, W, Hiraizumi, M, Shimada, H, Yamashita, K, Nishizawa, T, Nureki, O. | | 登録日 | 2019-12-26 | | 公開日 | 2020-07-29 | | 最終更新日 | 2020-09-09 | | 実験手法 | ELECTRON MICROSCOPY (2.66 Å) | | 主引用文献 | Cryo-EM structures of calcium homeostasis modulator channels in diverse oligomeric assemblies.
Sci Adv, 6, 2020
|
|
6LMW
 
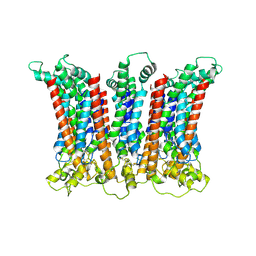 | | Cryo-EM structure of the CALHM chimeric construct (8-mer) | | 分子名称: | Calcium homeostasis modulator 1,Calcium homeostasis modulator protein 2 | | 著者 | Demura, K, Kusakizako, T, Shihoya, W, Hiraizumi, M, Shimada, H, Yamashita, K, Nishizawa, T, Nureki, O. | | 登録日 | 2019-12-26 | | 公開日 | 2020-07-29 | | 最終更新日 | 2020-09-09 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Cryo-EM structures of calcium homeostasis modulator channels in diverse oligomeric assemblies.
Sci Adv, 6, 2020
|
|
5G2X
 
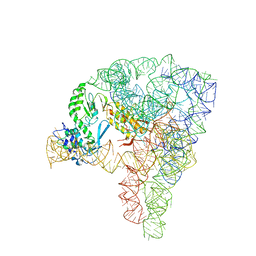 | | Structure a of Group II Intron Complexed with its Reverse Transcriptase | | 分子名称: | 5'-R(*CP*AP*CP*AP*UP*CP*CP*AP*UP*AP*AP*CP)-3', GROUP II INTRON, GROUP II INTRON-ENCODED PROTEIN LTRA | | 著者 | Qu, G, Kaushal, P.S, Wang, J, Shigematsu, H, Piazza, C.L, Agrawal, R.K, Belfort, M, Wang, H.W. | | 登録日 | 2016-04-16 | | 公開日 | 2016-05-11 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structure of a Group II Intron in Complex with its Reverse Transcriptase.
Nat.Struct.Mol.Biol., 23, 2016
|
|
6LMV
 
 | | Cryo-EM structure of the C. elegans CLHM-1 | | 分子名称: | Calcium homeostasis modulator protein | | 著者 | Demura, K, Kusakizako, T, Shihoya, W, Hiraizumi, M, Shimada, H, Yamashita, K, Nishizawa, T, Nureki, O. | | 登録日 | 2019-12-26 | | 公開日 | 2020-07-29 | | 最終更新日 | 2020-09-09 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Cryo-EM structures of calcium homeostasis modulator channels in diverse oligomeric assemblies.
Sci Adv, 6, 2020
|
|
6LMU
 
 | | Cryo-EM structure of the human CALHM2 | | 分子名称: | Calcium homeostasis modulator protein 2 | | 著者 | Demura, K, Kusakizako, T, Shihoya, W, Hiraizumi, M, Shimada, H, Yamashita, K, Nishizawa, T, Nureki, O. | | 登録日 | 2019-12-26 | | 公開日 | 2020-07-29 | | 最終更新日 | 2020-09-09 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Cryo-EM structures of calcium homeostasis modulator channels in diverse oligomeric assemblies.
Sci Adv, 6, 2020
|
|
6LMX
 
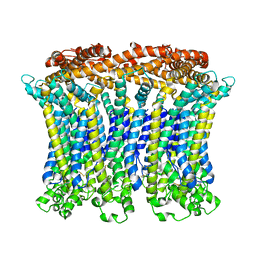 | | Cryo-EM structure of the CALHM chimeric construct (9-mer) | | 分子名称: | Calcium homeostasis modulator 1,Calcium homeostasis modulator protein 2 | | 著者 | Demura, K, Kusakizako, T, Shihoya, W, Hiraizumi, M, Shimada, H, Yamashita, K, Nishizawa, T, Nureki, O. | | 登録日 | 2019-12-26 | | 公開日 | 2020-07-29 | | 最終更新日 | 2020-09-09 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Cryo-EM structures of calcium homeostasis modulator channels in diverse oligomeric assemblies.
Sci Adv, 6, 2020
|
|
5G2Y
 
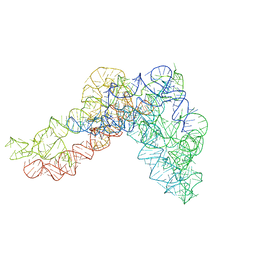 | | Structure a of Group II Intron Complexed with its Reverse Transcriptase | | 分子名称: | GROUP II INTRON | | 著者 | Qu, G, Kaushal, P.S, Wang, J, Shigematsu, H, Piazza, C.L, Agrawal, R.K, Belfort, M, Wang, H.W. | | 登録日 | 2016-04-16 | | 公開日 | 2016-05-04 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | Structure of a Group II Intron in Complex with its Reverse Transcriptase.
Nat.Struct.Mol.Biol., 23, 2016
|
|
2AHL
 
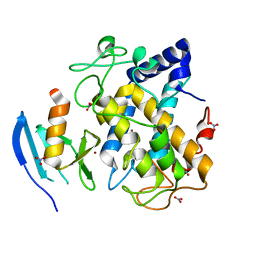 | | Crystal structure of the hydroxylamine-induced deoxy-form of the copper-bound Streptomyces castaneoglobisporus tyrosinase in complex with a caddie protein | | 分子名称: | CADDIE PROTEIN ORF378, COPPER (I) ION, NITRATE ION, ... | | 著者 | Matoba, Y, Kumagai, T, Yamamoto, A, Yoshitsu, H, Sugiyama, M. | | 登録日 | 2005-07-28 | | 公開日 | 2006-01-31 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Crystallographic Evidence That the Dinuclear Copper Center of Tyrosinase Is Flexible during Catalysis
J.Biol.Chem., 281, 2006
|
|
2AHK
 
 | | Crystal structure of the met-form of the copper-bound Streptomyces castaneoglobisporus tyrosinase in complex with a caddie protein obtained by soking in cupric sulfate for 6 months | | 分子名称: | CADDIE PROTEIN ORF378, COPPER (II) ION, NITRATE ION, ... | | 著者 | Matoba, Y, Kumagai, T, Yamamoto, A, Yoshitsu, H, Sugiyama, M. | | 登録日 | 2005-07-28 | | 公開日 | 2006-01-31 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.71 Å) | | 主引用文献 | Crystallographic Evidence That the Dinuclear Copper Center of Tyrosinase Is Flexible during Catalysis
J.Biol.Chem., 281, 2006
|
|
