6C3K
 
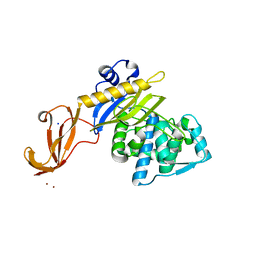 | |
6CO9
 
 | | Crystal structure of Rhodococcus jostii RHA1 IpdAB COCHEA-COA complex | | 分子名称: | Probable CoA-transferase alpha subunit, Probable CoA-transferase beta subunit, S-{(3R,5R,9R)-1-[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]-3,5,9-trihydroxy-8,8-dimethyl-3,5-dioxido-10,14-dioxo-2,4,6-trioxa-11,15-diaza-3lambda~5~,5lambda~5~-diphosphaheptadecan-17-yl} (5R,10R)-7-hydroxy-10-methyl-2-oxo-1-oxaspiro[4.5]dec-6-ene-6-carbothioate (non-preferred name), ... | | 著者 | Crowe, A.M, Workman, S.D, Watanabe, N, Worrall, L.J, Strynadka, N.C.J, Eltis, L.D. | | 登録日 | 2018-03-12 | | 公開日 | 2018-03-28 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.602 Å) | | 主引用文献 | IpdAB, a virulence factor inMycobacterium tuberculosis, is a cholesterol ring-cleaving hydrolase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
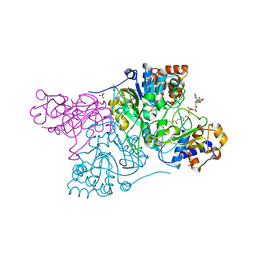
6CB2
 
 | | Crystal structure of Escherichia coli UppP | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, SULFATE ION, Undecaprenyl-diphosphatase | | 著者 | Workman, S.D, Worrall, L.J, Strynadka, N.C.J. | | 登録日 | 2018-02-01 | | 公開日 | 2018-03-28 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of an intramembranal phosphatase central to bacterial cell-wall peptidoglycan biosynthesis and lipid recycling.
Nat Commun, 9, 2018
|
|
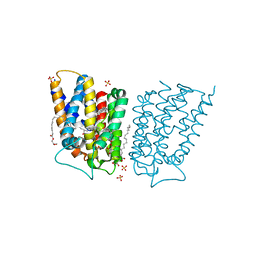
6COJ
 
 | | Crystal structure of Rhodococcus jostii RHA1 IpdAB E105A COCHEA-COA complex | | 分子名称: | Probable CoA-transferase alpha subunit, Probable CoA-transferase beta subunit, S-{(3R,5R,9R)-1-[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]-3,5,9-trihydroxy-8,8-dimethyl-3,5-dioxido-10,14-dioxo-2,4,6-trioxa-11,15-diaza-3lambda~5~,5lambda~5~-diphosphaheptadecan-17-yl} (5R,10R)-7-hydroxy-10-methyl-2-oxo-1-oxaspiro[4.5]dec-6-ene-6-carbothioate (non-preferred name), ... | | 著者 | Crowe, A.M, Workman, S.D, Watanabe, N, Worrall, L.J, Strynadka, N.C.J, Eltis, L.D. | | 登録日 | 2018-03-12 | | 公開日 | 2018-03-28 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | IpdAB, a virulence factor inMycobacterium tuberculosis, is a cholesterol ring-cleaving hydrolase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6C39
 
 | |
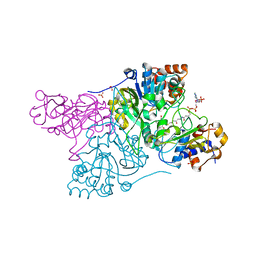
6CON
 
 | | Crystal structure of Mycobacterium tuberculosis IpdAB | | 分子名称: | CoA-transferase subunit alpha, CoA-transferase subunit beta | | 著者 | Crowe, A.M, Workman, S.D, Watanabe, N, Worrall, L.J, Strynadka, N.C.J, Eltis, L.D. | | 登録日 | 2018-03-12 | | 公開日 | 2018-03-28 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | IpdAB, a virulence factor inMycobacterium tuberculosis, is a cholesterol ring-cleaving hydrolase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6CO6
 
 | | Crystal structure of Rhodococcus jostii RHA1 IpdAB | | 分子名称: | GLYCEROL, Probable CoA-transferase alpha subunit, Probable CoA-transferase beta subunit, ... | | 著者 | Crowe, A.M, Workman, S.D, Watanabe, N, Worrall, L.J, Strynadka, N.C.J, Eltis, L.D. | | 登録日 | 2018-03-12 | | 公開日 | 2018-03-28 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.701 Å) | | 主引用文献 | IpdAB, a virulence factor inMycobacterium tuberculosis, is a cholesterol ring-cleaving hydrolase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
1Q16
 
 | | Crystal structure of Nitrate Reductase A, NarGHI, from Escherichia coli | | 分子名称: | (1S)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PENTANOYLOXY)METHYL]ETHYL OCTANOATE, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, FE3-S4 CLUSTER, ... | | 著者 | Bertero, M.G, Strynadka, N.C.J. | | 登録日 | 2003-07-18 | | 公開日 | 2003-10-07 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Insights into the respiratory electron transfer pathway from the structure of nitrate reductase A
Nat.Struct.Biol., 10, 2003
|
|
1Y5I
 
 | | The crystal structure of the NarGHI mutant NarI-K86A | | 分子名称: | (1S)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PENTANOYLOXY)METHYL]ETHYL OCTANOATE, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, FE3-S4 CLUSTER, ... | | 著者 | Bertero, M.G, Rothery, R.A, Boroumand, N, Palak, M, Blasco, F, Ginet, N, Weiner, J.H, Strynadka, N.C.J. | | 登録日 | 2004-12-02 | | 公開日 | 2005-03-08 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural and Biochemical Characterization of a Quinol Binding Site of Escherichia coli Nitrate Reductase A
J.Biol.Chem., 280, 2005
|
|
1XOU
 
 | |
1Y5L
 
 | | The crystal structure of the NarGHI mutant NarI-H66Y | | 分子名称: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, FE3-S4 CLUSTER, IRON/SULFUR CLUSTER, ... | | 著者 | Bertero, M.G, Rothery, R.A, Boroumand, N, Palak, M, Blasco, F, Ginet, N, Weiner, J.H, Strynadka, N.C.J. | | 登録日 | 2004-12-02 | | 公開日 | 2005-03-08 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural and Biochemical Characterization of a Quinol Binding Site of Escherichia coli Nitrate Reductase A
J.Biol.Chem., 280, 2005
|
|
1Y4Z
 
 | | The crystal structure of Nitrate Reductase A, NarGHI, in complex with the Q-site inhibitor pentachlorophenol | | 分子名称: | (1S)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PENTANOYLOXY)METHYL]ETHYL OCTANOATE, FE3-S4 CLUSTER, IRON/SULFUR CLUSTER, ... | | 著者 | Bertero, M.G, Rothery, R.A, Boroumand, N, Palak, M, Blasco, F, Ginet, N, Weiner, J.H, Strynadka, N.C.J. | | 登録日 | 2004-12-01 | | 公開日 | 2005-03-08 | | 最終更新日 | 2021-11-10 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural and Biochemical Characterization of a Quinol Binding Site of Escherichia coli Nitrate Reductase A
J.Biol.Chem., 280, 2005
|
|
1Y5N
 
 | | The crystal structure of the NarGHI mutant NarI-K86A in complex with pentachlorophenol | | 分子名称: | (1S)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PENTANOYLOXY)METHYL]ETHYL OCTANOATE, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, FE3-S4 CLUSTER, ... | | 著者 | Bertero, M.G, Rothery, R.A, Boroumand, N, Palak, M, Blasco, F, Ginet, N, Weiner, J.H, Strynadka, N.C.J. | | 登録日 | 2004-12-02 | | 公開日 | 2005-03-08 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural and Biochemical Characterization of a Quinol Binding Site of Escherichia coli Nitrate Reductase A
J.Biol.Chem., 280, 2005
|
|
3ECH
 
 | |
3EGW
 
 | | The crystal structure of the NarGHI mutant NarH - C16A | | 分子名称: | (1S)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PENTANOYLOXY)METHYL]ETHYL OCTANOATE, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, ... | | 著者 | Bertero, M.G, Rothery, R.A, Weiner, J.H, Strynadka, N.C.J. | | 登録日 | 2008-09-11 | | 公開日 | 2010-03-02 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | When width is more important than height: Barriers to electron transfer in E.coli nitrate reductase
To be Published
|
|
1K0W
 
 | | CRYSTAL STRUCTURE OF L-RIBULOSE-5-PHOSPHATE 4-EPIMERASE | | 分子名称: | L-RIBULOSE 5 PHOSPHATE 4-EPIMERASE, ZINC ION | | 著者 | Luo, Y, Samuel, J, Mosimann, S.C, Lee, J.E, Strynadka, N.C.J. | | 登録日 | 2001-09-21 | | 公開日 | 2003-01-28 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | The structure of L-ribulose-5-phosphate 4-epimerase: an aldolase-like platform for epimerization
Biochemistry, 40, 2001
|
|
1ERM
 
 | | X-RAY CRYSTAL STRUCTURE OF TEM-1 BETA LACTAMASE IN COMPLEX WITH A DESIGNED BORONIC ACID INHIBITOR (1R)-1-ACETAMIDO-2-(3-CARBOXYPHENYL)ETHANE BORONIC ACID | | 分子名称: | 1(R)-1-ACETAMIDO-2-(3-CARBOXYPHENYL)ETHYL BORONIC ACID, TEM-1 BETA-LACTAMASE | | 著者 | Ness, S, Martin, R, Kindler, A.M, Paetzel, M, Gold, M, Jones, J.B, Strynadka, N.C.J. | | 登録日 | 2000-04-06 | | 公開日 | 2000-05-10 | | 最終更新日 | 2012-06-20 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structure-based design guides the improved efficacy of deacylation transition state analogue inhibitors of TEM-1 beta-Lactamase(,).
Biochemistry, 39, 2000
|
|
8SXR
 
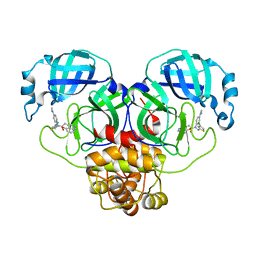 | | Crystal structure of SARS-CoV-2 Mpro with C5a | | 分子名称: | 3C-like proteinase nsp5, N-[(4-chlorothiophen-2-yl)methyl]-N-[4-(dimethylamino)phenyl]-2-(5-hydroxyisoquinolin-4-yl)acetamide | | 著者 | Worrall, L.J, Kenward, C, Lee, J, Strynadka, N.C.J. | | 登録日 | 2023-05-23 | | 公開日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.114 Å) | | 主引用文献 | A novel class of broad-spectrum active-site-directed 3C-like protease inhibitors with nanomolar antiviral activity against highly immune-evasive SARS-CoV-2 Omicron subvariants.
Emerg Microbes Infect, 12, 2023
|
|
8TDH
 
 | |
8TDE
 
 | |
8TDA
 
 | |
8TDF
 
 | |
8TCT
 
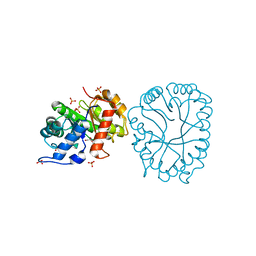 | | Structure of 3K-GlcH bound Bacteroides thetaiotaomicron 3-Keto-beta-glucopyranoside-1,2-Lyase BT1 | | 分子名称: | 1,5-anhydro-D-ribo-hex-3-ulose, COBALT (II) ION, PHOSPHATE ION, ... | | 著者 | Lazarski, A.C, Worrall, L.J, Strynadka, N.C.J. | | 登録日 | 2023-07-02 | | 公開日 | 2024-06-12 | | 最終更新日 | 2024-07-03 | | 実験手法 | X-RAY DIFFRACTION (1.86 Å) | | 主引用文献 | An alternative broad-specificity pathway for glycan breakdown in bacteria.
Nature, 2024
|
|
8TCD
 
 | | Structure of Alistipes sp. 3-Keto-beta-glucopyranoside-1,2-Lyase AL1 | | 分子名称: | ACETATE ION, COBALT (II) ION, GLYCEROL, ... | | 著者 | Lazarski, A.C, Worrall, L.J, Strynadka, N.C.J. | | 登録日 | 2023-06-30 | | 公開日 | 2024-06-12 | | 最終更新日 | 2024-07-03 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | An alternative broad-specificity pathway for glycan breakdown in bacteria.
Nature, 2024
|
|
8TDI
 
 | | Structure of P2B11 Glucuronide-3-dehydrogenase | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, P2B11 Glucuronide-3-dehydrogenase, ... | | 著者 | Lazarski, A.C, Worrall, L.J, Strynadka, N.C.J. | | 登録日 | 2023-07-03 | | 公開日 | 2024-06-12 | | 最終更新日 | 2024-07-03 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | An alternative broad-specificity pathway for glycan breakdown in bacteria.
Nature, 2024
|
|
