5LE4
 
 | | Crystal structure of DARPin-DARPin rigid fusion, variant DD_D12_11_D12 | | 分子名称: | DD_D12_11_D12 | | 著者 | Batyuk, A, Wu, Y, Mittl, P.R, Plueckthun, A. | | 登録日 | 2016-06-29 | | 公開日 | 2017-08-02 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Rigidly connected multispecific artificial binders with adjustable geometries.
Sci Rep, 7, 2017
|
|
5LEA
 
 | | Crystal structure of DARPin-DARPin rigid fusion, variant DD_Off7_12_3G124 | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, DD_Off7_12_3G124 | | 著者 | Batyuk, A, Wu, Y, Mittl, P.R, Plueckthun, A. | | 登録日 | 2016-06-29 | | 公開日 | 2017-08-02 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Rigidly connected multispecific artificial binders with adjustable geometries.
Sci Rep, 7, 2017
|
|
5LED
 
 | | Crystal structure of DARPin-DARPin rigid fusion, variant DDD_D12_12_D12_12_D12 | | 分子名称: | DDD_D12_12_D12_12_D12 | | 著者 | Batyuk, A, Wu, Y, Mittl, P.R, Plueckthun, A. | | 登録日 | 2016-06-29 | | 公開日 | 2017-08-02 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Rigidly connected multispecific artificial binders with adjustable geometries.
Sci Rep, 7, 2017
|
|
5LEM
 
 | | Crystal structure of DARPin-DARPin rigid fusion, variant DD_Off7_11_3G124 in complex with Maltose-binding Protein and Green Fluorescent Protein | | 分子名称: | DD_Off7_11_3G124, Green fluorescent protein, Maltose-binding periplasmic protein | | 著者 | Batyuk, A, Wu, Y, Mittl, P.R, Plueckthun, A. | | 登録日 | 2016-06-30 | | 公開日 | 2017-08-02 | | 最終更新日 | 2019-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.98 Å) | | 主引用文献 | Rigidly connected multispecific artificial binders with adjustable geometries.
Sci Rep, 7, 2017
|
|
5LE6
 
 | | Crystal structure of DARPin-DARPin rigid fusion, variant DD_D12_09_D12 | | 分子名称: | DD_D12_09_D12, GLYCEROL, SULFATE ION | | 著者 | Batyuk, A, Wu, Y, Mittl, P.R, Plueckthun, A. | | 登録日 | 2016-06-29 | | 公開日 | 2017-08-02 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Rigidly connected multispecific artificial binders with adjustable geometries.
Sci Rep, 7, 2017
|
|
7AQH
 
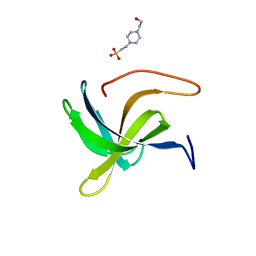 | | Cell wall binding domain of the Staphylococcal phage 2638A endolysin | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ORF007 | | 著者 | Dunne, M, Sobieraj, A, Ernst, P, Mittl, P.R.E, Pluckthun, A, Loessner, M.J. | | 登録日 | 2020-10-21 | | 公開日 | 2020-12-02 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.49 Å) | | 主引用文献 | Cell wall binding domain of the Staphylococcal phage 2638A endolysin
To Be Published
|
|
8QBC
 
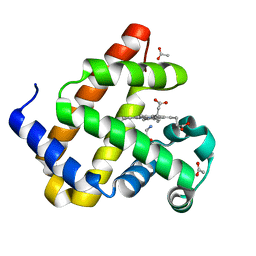 | | Reactive amide intermediate in sperm whale myoglobin mutant H64V V68A | | 分子名称: | ACETATE ION, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | 著者 | Tinzl, M, Mittl, P.R.E, Hilvert, D. | | 登録日 | 2023-08-24 | | 公開日 | 2023-12-13 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.23 Å) | | 主引用文献 | Myoglobin-Catalyzed Azide Reduction Proceeds via an Anionic Metal Amide Intermediate.
J.Am.Chem.Soc., 146, 2024
|
|
8QBA
 
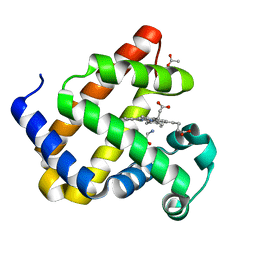 | | Sperm whale myoblogin mutant H64V V68A in complex with glycine ethyl ester | | 分子名称: | ACETATE ION, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | 著者 | Tinzl, M, Mittl, P.R.E, Hilvert, D. | | 登録日 | 2023-08-24 | | 公開日 | 2023-12-13 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.39 Å) | | 主引用文献 | Myoglobin-Catalyzed Azide Reduction Proceeds via an Anionic Metal Amide Intermediate.
J.Am.Chem.Soc., 146, 2024
|
|
2CJY
 
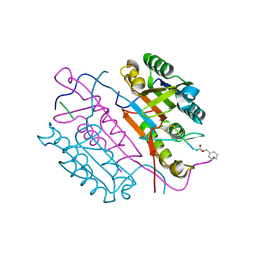 | | Extended substrate recognition in caspase-3 revealed by high resolution X-ray structure analysis | | 分子名称: | CASPASE-3, PHQ-ASP-GLU-VAL-ASP-CHLOROMETHYLKETONE | | 著者 | Ganesan, R, Mittl, P.R.E, Jelakovic, S, Grutter, M.G. | | 登録日 | 2006-04-09 | | 公開日 | 2006-06-27 | | 最終更新日 | 2017-06-28 | | 実験手法 | X-RAY DIFFRACTION (1.67 Å) | | 主引用文献 | Extended Substrate Recognition in Caspase-3 Revealed by High Resolution X-Ray Structure Analysis
J.Mol.Biol., 359, 2006
|
|
7QNP
 
 | | Designed Armadillo repeat protein N(A4)M4C(AII) co-crystallized with hen egg white lysozyme | | 分子名称: | 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, Designed Armadillo Repeat Protein N(A4)M4C(AII), ... | | 著者 | Michel, E, Mittl, P.R.E, Plueckthun, A. | | 登録日 | 2021-12-21 | | 公開日 | 2022-06-08 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.586 Å) | | 主引用文献 | Improved Repeat Protein Stability by Combined Consensus and Computational Protein Design.
Biochemistry, 62, 2023
|
|
2CJX
 
 | | Extended substrate recognition in caspase-3 revealed by high resolution X-ray structure analysis | | 分子名称: | CASPASE-3, PHQ-ASP-GLU-VAL-ASP-CHLOROMETHYLKETONE | | 著者 | Ganesan, R, Mittl, P.R.E, Jelakovic, S, Grutter, M.G. | | 登録日 | 2006-04-09 | | 公開日 | 2006-06-27 | | 最終更新日 | 2017-06-28 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Extended Substrate Recognition in Caspase-3 Revealed by High Resolution X-Ray Structure Analysis
J.Mol.Biol., 359, 2006
|
|
6QFO
 
 | | EngBF DARPin Fusion 9b 3G124 | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MANGANESE (II) ION, ... | | 著者 | Ernst, P, Pluckthun, A, Mittl, P.R.E. | | 登録日 | 2019-01-10 | | 公開日 | 2019-11-06 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural analysis of biological targets by host:guest crystal lattice engineering.
Sci Rep, 9, 2019
|
|
6QEP
 
 | | EngBF DARPin Fusion 4b H14 | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MANGANESE (II) ION, ... | | 著者 | Ernst, P, Pluckthun, A, Mittl, P.R.E. | | 登録日 | 2019-01-08 | | 公開日 | 2019-11-06 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structural analysis of biological targets by host:guest crystal lattice engineering.
Sci Rep, 9, 2019
|
|
6QEV
 
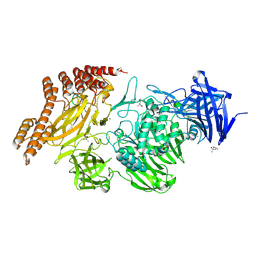 | | EngBF DARPin Fusion 4b B6 | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, MANGANESE (II) ION, PEGA domain-containing protein,PEGA domain-containing protein,EngBF DARPin fusion B6 complex, ... | | 著者 | Ernst, P, Pluckthun, A, Mittl, P.R.E. | | 登録日 | 2019-01-08 | | 公開日 | 2019-11-06 | | 最終更新日 | 2020-04-22 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structural analysis of biological targets by host:guest crystal lattice engineering.
Sci Rep, 9, 2019
|
|
6QFK
 
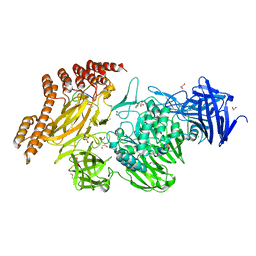 | | EngBF DARPin Fusion 4b G10 | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | 著者 | Ernst, P, Pluckthun, A, Mittl, P.R.E. | | 登録日 | 2019-01-10 | | 公開日 | 2019-11-06 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural analysis of biological targets by host:guest crystal lattice engineering.
Sci Rep, 9, 2019
|
|
6SA7
 
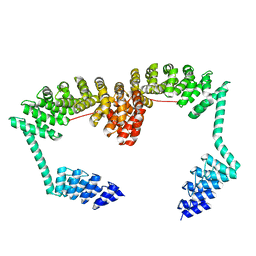 | | DARPin-Armadillo fusion C8long83 | | 分子名称: | DARPin-Armadillo fusion C8long83 | | 著者 | Ernst, P, Honegger, A, van der Valk, F, Ewald, C, Mittl, P.R.E, Plucktun, A. | | 登録日 | 2019-07-16 | | 公開日 | 2019-11-20 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Rigid fusions of designed helical repeat binding proteins efficiently protect a binding surface from crystal contacts.
Sci Rep, 9, 2019
|
|
6SA8
 
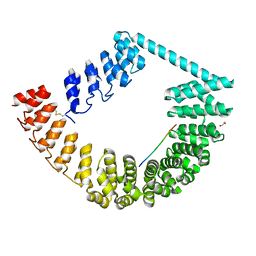 | | ring-like DARPin-Armadillo fusion H83_D01 | | 分子名称: | 1,2-ETHANEDIOL, LYS-ARG-LYS-ARG-LYS-ARG-LYS-ARG-LYS-ARG, ring-like DARPin-Armadillo fusion H83_D01 | | 著者 | Ernst, P, Honegger, A, van der Valk, F, Ewald, C, Mittl, P.R.E, Plucktun, A. | | 登録日 | 2019-07-16 | | 公開日 | 2019-11-20 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Rigid fusions of designed helical repeat binding proteins efficiently protect a binding surface from crystal contacts.
Sci Rep, 9, 2019
|
|
6SH9
 
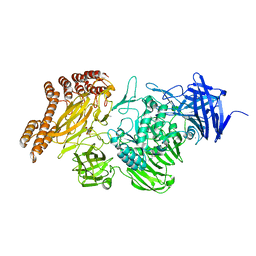 | | EngBF DARPin Fusion 4b D12 | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Endo-alpha-N-acetylgalactosaminidase,DARPin 4b D12, ... | | 著者 | Ernst, P, Pluckthun, A, Mittl, P.R.E. | | 登録日 | 2019-08-06 | | 公開日 | 2019-11-06 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural analysis of biological targets by host:guest crystal lattice engineering.
Sci Rep, 9, 2019
|
|
6SA6
 
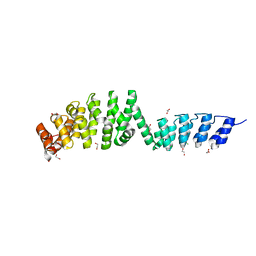 | | DARPin-Armadillo fusion A5 | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, DARPin-Armadillo fusion A5 | | 著者 | Ernst, P, Honegger, A, van der Valk, F, Ewald, C, Mittl, P.R.E, Pluckthun, A. | | 登録日 | 2019-07-16 | | 公開日 | 2019-11-20 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Rigid fusions of designed helical repeat binding proteins efficiently protect a binding surface from crystal contacts.
Sci Rep, 9, 2019
|
|
6Z4T
 
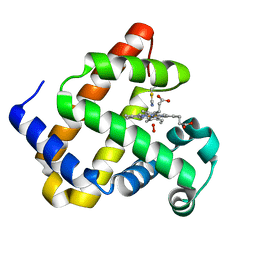 | |
6Z4R
 
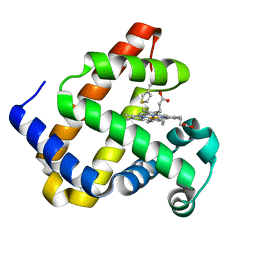 | |
4BS0
 
 | | Crystal Structure of Kemp Eliminase HG3.17 E47N,N300D Complexed with Transition State Analog 6-Nitrobenzotriazole | | 分子名称: | 6-NITROBENZOTRIAZOLE, KEMP ELIMINASE HG3.17, SULFATE ION | | 著者 | Blomberg, R, Kries, H, Pinkas, D.M, Mittl, P.R.E, Gruetter, M.G, Privett, H.K, Mayo, S, Hilvert, D. | | 登録日 | 2013-06-06 | | 公開日 | 2013-10-16 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.09 Å) | | 主引用文献 | Precision is Essential for Efficient Catalysis in an Evolved Kemp Eliminase
Nature, 503, 2013
|
|
4CG4
 
 | | Crystal structure of the CHS-B30.2 domains of TRIM20 | | 分子名称: | 1,2-ETHANEDIOL, 2-AMINO-ETHANETHIOL, PYRIN, ... | | 著者 | Weinert, C, Morger, D, Djekic, A, Mittl, P.R.E, Gruetter, M.G. | | 登録日 | 2013-11-20 | | 公開日 | 2015-04-29 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal Structure of Trim20 C-Terminal Coiled-Coil/B30.2 Fragment: Implications for the Recognition of Higher Order Oligomers
Sci.Rep., 5, 2015
|
|
4D49
 
 | | Crystal structure of computationally designed armadillo repeat proteins for modular peptide recognition. | | 分子名称: | ARGININE, ARMADILLO REPEAT PROTEIN ARM00027, POLY ARG DECAPEPTIDE | | 著者 | Reichen, C, Forzani, C, Zhou, T, Parmeggiani, F, Fleishman, S.J, Mittl, P.R.E, Madhurantakam, C, Honegger, A, Ewald, C, Zerbe, O, Baker, D, Caflisch, A, Pluckthun, A. | | 登録日 | 2014-10-27 | | 公開日 | 2016-01-13 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.09 Å) | | 主引用文献 | Computationally Designed Armadillo Repeat Proteins for Modular Peptide Recognition.
J.Mol.Biol., 428, 2016
|
|
4D4E
 
 | | Crystal structure of computationally designed armadillo repeat proteins for modular peptide recognition. | | 分子名称: | ARMADILLO REPEAT PROTEIN ARM00016, GLYCEROL | | 著者 | Reichen, C, Forzani, C, Zhou, T, Parmeggiani, F, Fleishman, S.J, Mittl, P.R.E, Madhurantakam, C, Honegger, A, Ewald, C, Zerbe, O, Baker, D, Caflisch, A, Pluckthun, A. | | 登録日 | 2014-10-28 | | 公開日 | 2016-01-13 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Computationally Designed Armadillo Repeat Proteins for Modular Peptide Recognition.
J.Mol.Biol., 428, 2016
|
|
