2SXL
 
 | | SEX-LETHAL RBD1, NMR, MINIMIZED AVERAGE STRUCTURE | | 分子名称: | SEX-LETHAL PROTEIN | | 著者 | Inoue, M, Muto, Y, Sakamoto, H, Kigawa, T, Takio, K, Shimura, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 1997-07-16 | | 公開日 | 1998-07-22 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | A characteristic arrangement of aromatic amino acid residues in the solution structure of the amino-terminal RNA-binding domain of Drosophila sex-lethal.
J.Mol.Biol., 272, 1997
|
|
6BSR
 
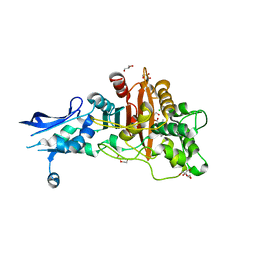 | | Crystal structure of penicillin-binding protein 4 (PBP4) from Enterococcus faecalis in the benzylpenicillin bound form. | | 分子名称: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | 著者 | Moon, T.M, D'Andrea, E.D, Peti, W, Page, R. | | 登録日 | 2017-12-04 | | 公開日 | 2018-10-31 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.34 Å) | | 主引用文献 | The structures of penicillin-binding protein 4 (PBP4) and PBP5 fromEnterococciprovide structural insights into beta-lactam resistance.
J. Biol. Chem., 293, 2018
|
|
6BSQ
 
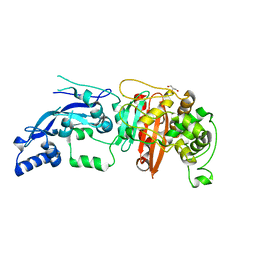 | | Enterococcus faecalis Penicillin Binding Protein 4 (PBP4) | | 分子名称: | CHLORIDE ION, GLYCEROL, PBP4 protein | | 著者 | Moon, T.M, D'Andrea, E.D, Peti, W, Page, R. | | 登録日 | 2017-12-04 | | 公開日 | 2018-10-31 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | The structures of penicillin-binding protein 4 (PBP4) and PBP5 fromEnterococciprovide structural insights into beta-lactam resistance.
J. Biol. Chem., 293, 2018
|
|
1C2X
 
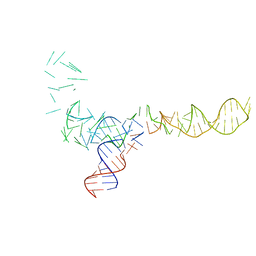 | |
1C2W
 
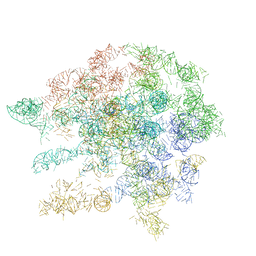 | |
1MNX
 
 | |
1SXL
 
 | |
1YB6
 
 | |
1YB7
 
 | | Hydroxynitrile lyase from hevea brasiliensis in complex with 2,3-dimethyl-2-hydroxy-butyronitrile | | 分子名称: | (S)-2-HYDROXY-2,3-DIMETHYLBUTANENITRILE, (S)-acetone-cyanohydrin lyase, SULFATE ION | | 著者 | Gruber, K, Gartler, G, Kratky, C. | | 登録日 | 2004-12-20 | | 公開日 | 2005-12-20 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | Structural determinants of the enantioselectivity of the hydroxynitrile lyase from Hevea brasiliensis
J.Biotechnol., 129, 2007
|
|
1SCI
 
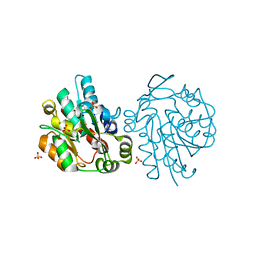 | | K236L mutant of hydroxynitrile lyase from Hevea brasiliensis | | 分子名称: | (S)-acetone-cyanohydrin lyase, SULFATE ION | | 著者 | Gruber, K, Gartler, G, Krammer, B, Schwab, H, Kratky, C. | | 登録日 | 2004-02-12 | | 公開日 | 2004-06-29 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.18 Å) | | 主引用文献 | Reaction mechanism of hydroxynitrile lyases of the alpha/beta-hydrolase superfamily: the three-dimensional structure of the transient enzyme-substrate complex certifies the crucial role of LYS236
J.Biol.Chem., 279, 2004
|
|
1SCK
 
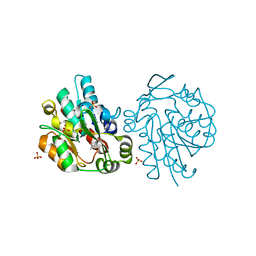 | | K236L mutant of hydroxynitrile lyase from Hevea brasiliensis in complex with acetone | | 分子名称: | (S)-acetone-cyanohydrin lyase, ACETONE, SULFATE ION | | 著者 | Gruber, K, Gartler, G, Krammer, B, Schwab, H, Kratky, C. | | 登録日 | 2004-02-12 | | 公開日 | 2004-06-29 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Reaction mechanism of hydroxynitrile lyases of the alpha/beta-hydrolase superfamily: the three-dimensional structure of the transient enzyme-substrate complex certifies the crucial role of LYS236
J.Biol.Chem., 279, 2004
|
|
1SCQ
 
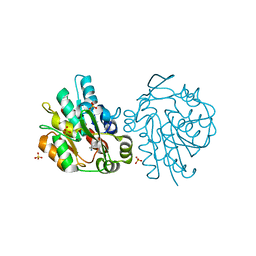 | | K236L mutant of hydroxynitrile lyase from Hevea brasiliensis in complex with acetonecyanohydrin | | 分子名称: | (S)-acetone-cyanohydrin lyase, 2-HYDROXY-2-METHYLPROPANENITRILE, SULFATE ION | | 著者 | Gruber, K, Gartler, G, Krammer, B, Schwab, H, Kratky, C. | | 登録日 | 2004-02-12 | | 公開日 | 2004-06-29 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Reaction mechanism of hydroxynitrile lyases of the alpha/beta-hydrolase superfamily: the three-dimensional structure of the transient enzyme-substrate complex certifies the crucial role of LYS236
J.Biol.Chem., 279, 2004
|
|
1SC9
 
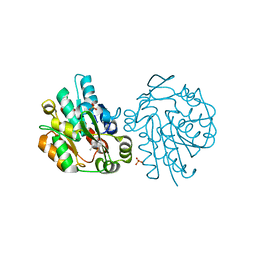 | | Hydroxynitrile Lyase from Hevea brasiliensis in complex with the natural substrate acetone cyanohydrin | | 分子名称: | (S)-acetone-cyanohydrin lyase, 2-HYDROXY-2-METHYLPROPANENITRILE, SULFATE ION | | 著者 | Gruber, K, Gartler, G, Krammer, B, Schwab, H, Kratky, C. | | 登録日 | 2004-02-12 | | 公開日 | 2004-06-29 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Reaction mechanism of hydroxynitrile lyases of the alpha/beta-hydrolase superfamily: the three-dimensional structure of the transient enzyme-substrate complex certifies the crucial role of LYS236
J.Biol.Chem., 279, 2004
|
|
8C8H
 
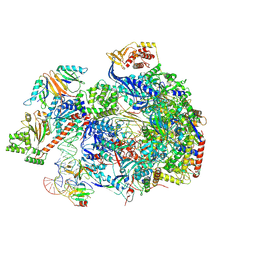 | |
6RIC
 
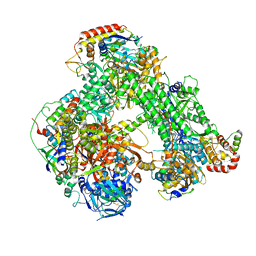 | | Structure of the core Vaccinia Virus DNA-dependent RNA polymerase complex | | 分子名称: | DNA-dependent RNA polymerase subunit rpo132, DNA-dependent RNA polymerase subunit rpo147, DNA-dependent RNA polymerase subunit rpo18, ... | | 著者 | Grimm, C, Hillen, H.S, Bedenk, K, Bartuli, J, Neyer, S, Zhang, Q, Huettenhofer, A, Erlacher, M, Dienemann, C, Schlosser, A, Urlaub, H, Boettcher, B, Szalay, A, Cramer, P, Fischer, U. | | 登録日 | 2019-04-23 | | 公開日 | 2019-12-18 | | 最終更新日 | 2019-12-25 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Structural Basis of Poxvirus Transcription: Transcribing and Capping Vaccinia Complexes.
Cell, 179, 2019
|
|
2LJJ
 
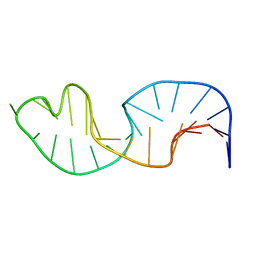 | |
