8DOY
 
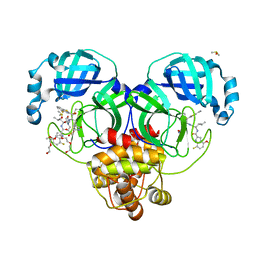 | | Crystal structure of SARS-CoV-2 main protease in complex with an inhibitor TKB-198 | | 分子名称: | 2-(2-METHOXYETHOXY)ETHANOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3C-like proteinase nsp5, ... | | 著者 | Bulut, H, Hayashi, H, Tsuji, K, Kuwata, N, Das, D, Tamamura, H, Mitsuya, H. | | 登録日 | 2022-07-14 | | 公開日 | 2022-08-24 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.59 Å) | | 主引用文献 | Potent and biostable inhibitors of the main protease of SARS-CoV-2.
Iscience, 25, 2022
|
|
1GZB
 
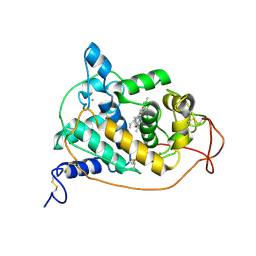 | | PEROXIDASE | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, PEROXIDASE, ... | | 著者 | Fukuyama, K, Kunishima, N, Amada, F. | | 登録日 | 1996-11-13 | | 公開日 | 1997-03-12 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Pentacoordination of the heme iron of Arthromyces ramosus peroxidase shown by a 1.8 A resolution crystallographic study at pH 4.5.
FEBS Lett., 378, 1996
|
|
6MKL
 
 | | X-ray crystal structure of darunavir-resistant-P51 HIV-1 protease in complex with GRL-142 | | 分子名称: | (3S,3aR,5R,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl [(2S,3R)-4-[{[2-(cyclopropylamino)-1,3-benzothiazol-6-yl]sulfonyl}(2-methylpropyl)amino]-1-(3,5-difluorophenyl)-3-hydroxybutan-2-yl]carbamate, Protease | | 著者 | Yedidi, R.S, Hayashi, H, Das, D, Mitsuya, H. | | 登録日 | 2018-09-25 | | 公開日 | 2019-10-02 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structure-function analysis of darunavir-resistant-P51 HIV-1 protease in complex with GRL-142.
To Be Published
|
|
7DFS
 
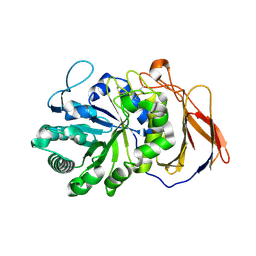 | | Crystal structure of a novel 4-O-alpha-L-rhamnosyl-beta-D-glucuronidase from Fusarium oxysporum 12S - Rha-GlcA complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-O-alpha-L-rhamnosyl-beta-D-glucuronidase, alpha-D-mannopyranose, ... | | 著者 | Kondo, T, Arakawa, T, Fushinobu, S, Sakamoto, T. | | 登録日 | 2020-11-09 | | 公開日 | 2021-03-17 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.49 Å) | | 主引用文献 | Biochemical and structural characterization of a novel 4-O-alpha-l-rhamnosyl-beta-d-glucuronidase from Fusarium oxysporum.
Febs J., 288, 2021
|
|
7DFQ
 
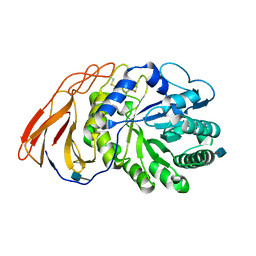 | | Crystal Structure of a novel 4-O-alpha-L-rhamnosyl-beta-D-glucuronidase from Fusarium oxysporum 12S, ligand-free form | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-O-alpha-L-rhamnosyl-beta-D-glucuronidase | | 著者 | Kondo, T, Arakawa, T, Fushinobu, S, Sakamoto, T. | | 登録日 | 2020-11-09 | | 公開日 | 2021-03-17 | | 最終更新日 | 2021-08-25 | | 実験手法 | X-RAY DIFFRACTION (1.51 Å) | | 主引用文献 | Biochemical and structural characterization of a novel 4-O-alpha-l-rhamnosyl-beta-d-glucuronidase from Fusarium oxysporum.
Febs J., 288, 2021
|
|
1GZA
 
 | | PEROXIDASE | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, IODIDE ION, ... | | 著者 | Fukuyama, K, Itakura, H. | | 登録日 | 1996-11-13 | | 公開日 | 1997-03-12 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.06 Å) | | 主引用文献 | Binding of iodide to Arthromyces ramosus peroxidase investigated with X-ray crystallographic analysis, 1H and 127I NMR spectroscopy, and steady-state kinetics.
J.Biol.Chem., 272, 1997
|
|
1HSR
 
 | | BINDING MODE OF BENZHYDROXAMIC ACID TO ARTHROMYCES RAMOSUS PEROXIDASE | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BENZHYDROXAMIC ACID, CALCIUM ION, ... | | 著者 | Fukuyama, K, Itakura, H. | | 登録日 | 1997-07-01 | | 公開日 | 1998-07-01 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Binding mode of benzhydroxamic acid to Arthromyces ramosus peroxidase shown by X-ray crystallographic analysis of the complex at 1.6 A resolution.
FEBS Lett., 412, 1997
|
|
7ESM
 
 | | Crystal structure of a L-rhamnose-alpha-1,4-D-glucuronate lyase from Fusarium oxysporum 12S, L-Rha complex | | 分子名称: | ACETATE ION, L-rhamnose-alpha-1,4-D-glucuronate lyase, SODIUM ION, ... | | 著者 | Kondo, T, Arakawa, T, Fushinobu, S, Sakamoto, T. | | 登録日 | 2021-05-11 | | 公開日 | 2021-08-04 | | 最終更新日 | 2021-09-01 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Structural and functional analysis of gum arabic l-rhamnose-alpha-1,4-d-glucuronate lyase establishes a novel polysaccharide lyase family.
J.Biol.Chem., 297, 2021
|
|
7ESN
 
 | | Crystal structure of a L-rhamnose-alpha-1,4-D-glucuronate lyase from Fusarium oxysporum 12S, H105F Rha-GlcA complex | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, L-Rhamnose-alpha-1,4-D-glucuronate lyase, ... | | 著者 | Kondo, T, Arakawa, T, Fushinobu, S, Sakamoto, T. | | 登録日 | 2021-05-11 | | 公開日 | 2021-08-04 | | 最終更新日 | 2021-09-01 | | 実験手法 | X-RAY DIFFRACTION (2.42 Å) | | 主引用文献 | Structural and functional analysis of gum arabic l-rhamnose-alpha-1,4-d-glucuronate lyase establishes a novel polysaccharide lyase family.
J.Biol.Chem., 297, 2021
|
|
7ESK
 
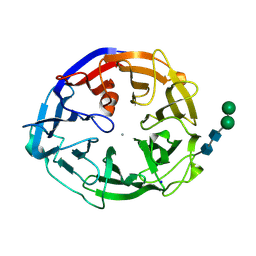 | | Crystal structure of a L-rhamnose-alpha-1,4-D-glucuronate lyase from Fusarium oxysporum 12S, Ligand free form | | 分子名称: | CALCIUM ION, L-Rhamnose-alpha-1,4-D-glucuronate lyase, SODIUM ION, ... | | 著者 | Kondo, T, Arakawa, T, Fushinobu, S, Sakamoto, T. | | 登録日 | 2021-05-11 | | 公開日 | 2021-08-04 | | 最終更新日 | 2021-09-01 | | 実験手法 | X-RAY DIFFRACTION (1.05 Å) | | 主引用文献 | Structural and functional analysis of gum arabic l-rhamnose-alpha-1,4-d-glucuronate lyase establishes a novel polysaccharide lyase family.
J.Biol.Chem., 297, 2021
|
|
7ESL
 
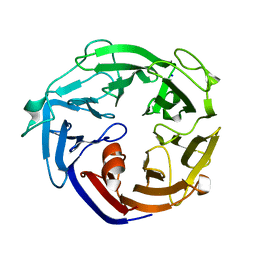 | | Crystal structure of a L-rhamnose-alpha-1,4-D-glucuronate lyase from Fusarium oxysporum 12S, N247A N-glycan free form | | 分子名称: | L-rhamnose-alpha-1,4-D-glucuronate lyase, SODIUM ION | | 著者 | Kondo, T, Arakawa, T, Fushinobu, S, Sakamoto, T. | | 登録日 | 2021-05-11 | | 公開日 | 2021-08-04 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Structural and functional analysis of gum arabic l-rhamnose-alpha-1,4-d-glucuronate lyase establishes a novel polysaccharide lyase family.
J.Biol.Chem., 297, 2021
|
|
1IXD
 
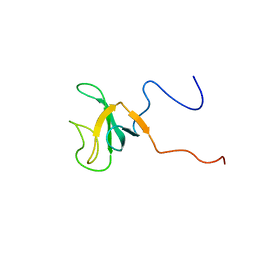 | | Solution structure of the CAP-GLY domain from human cylindromatosis tomour-suppressor CYLD | | 分子名称: | Cylindromatosis tumour-suppressor CYLD | | 著者 | Saito, K, Koshiba, S, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2002-06-19 | | 公開日 | 2002-12-19 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The CAP-Gly domain of CYLD associates with the proline-rich sequence in NEMO/IKKgamma
STRUCTURE, 12, 2004
|
|
8TH2
 
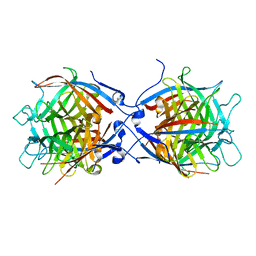 | | Structure of the isoflavene-forming dirigent protein PsPTS2 | | 分子名称: | Dirigent protein | | 著者 | Smith, C.A, Meng, Q, Lewis, N.G, Davin, L.B. | | 登録日 | 2023-07-13 | | 公開日 | 2024-01-31 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Dirigent isoflavene-forming PsPTS2: 3D structure, stereochemical, and kinetic characterization comparison with pterocarpan-forming PsPTS1 homolog in pea.
J.Biol.Chem., 300, 2024
|
|
1C8I
 
 | | BINDING MODE OF HYDROXYLAMINE TO ARTHROMYCES RAMOSUS PEROXIDASE | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, HYDROXYAMINE, ... | | 著者 | Wariishi, H, Nonaka, D, Johjima, T, Nakamura, N, Naruta, Y, Kubo, K, Fukuyama, K. | | 登録日 | 2000-05-08 | | 公開日 | 2001-01-17 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Direct binding of hydroxylamine to the heme iron of Arthromyces ramosus peroxidase. Substrate analogue that inhibits compound I formation in a competetive manner.
J.Biol.Chem., 275, 2000
|
|
1CK6
 
 | | BINDING MODE OF SALICYLHYDROXAMIC ACID TO ARTHROMYCES RAMOSUS PEROXIDASE | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, PROTEIN (PEROXIDASE), ... | | 著者 | Fukuyama, K, Itakura, H. | | 登録日 | 1999-04-28 | | 公開日 | 1999-12-29 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Binding of salicylhydroxamic acid and several aromatic donor molecules to Arthromyces ramosus peroxidase, investigated by X-ray crystallography, optical difference spectroscopy, NMR relaxation, molecular dynamics, and kinetics.
Biochemistry, 38, 1999
|
|
5ZRU
 
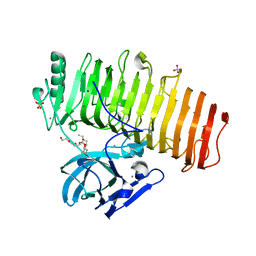 | | Crystal structure of Agl-KA catalytic domain | | 分子名称: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, Alpha-1,3-glucanase, CALCIUM ION, ... | | 著者 | Yano, S, Makabe, K. | | 登録日 | 2018-04-25 | | 公開日 | 2019-10-23 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.833 Å) | | 主引用文献 | Crystal structure of the catalytic unit of GH 87-type alpha-1,3-glucanase Agl-KA from Bacillus circulans.
Sci Rep, 9, 2019
|
|
3OCH
 
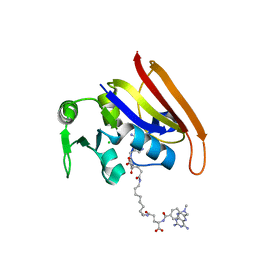 | | Chemically Self-assembled Antibody Nanorings (CSANs): Design and Characterization of an Anti-CD3 IgM Biomimetic | | 分子名称: | (2S,2'S)-5,5'-(nonane-1,9-diyldiimino)bis(2-{[(4-{[(2,4-diaminopteridin-6-yl)methyl](methyl)amino}phenyl)carbonyl]amino}-5-oxopentanoic acid) (non-preferred name), CHLORIDE ION, Dihydrofolate reductase, ... | | 著者 | Cody, V. | | 登録日 | 2010-08-10 | | 公開日 | 2010-12-08 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Chemically Self-Assembled Antibody Nanorings (CSANs): Design and Characterization of an Anti-CD3 IgM Biomimetic.
J.Am.Chem.Soc., 132, 2010
|
|
3IHA
 
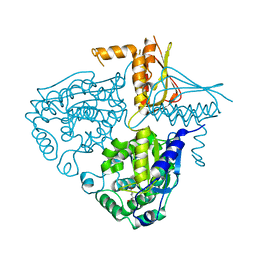 | |
3IH8
 
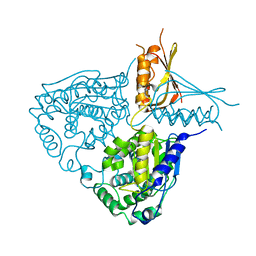 | |
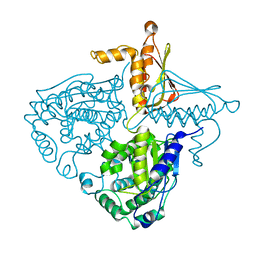
3IH9
 
 | |
3IHB
 
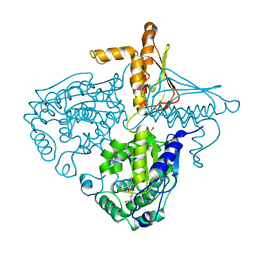 | |
1ARU
 
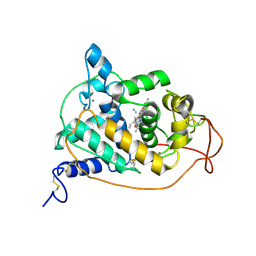 | | CRYSTAL STRUCTURES OF CYANIDE-AND TRIIODIDE-BOUND FORMS OF ARTHROMYCES RAMOSUS PEROXIDASE AT DIFFERENT PH VALUES. PERTURBATIONS OF ACTIVE SITE RESIDUES AND THEIR IMPLICATION IN ENZYME CATALYSIS | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CYANIDE ION, ... | | 著者 | Fukuyama, K, Kunishima, N, Amada, F. | | 登録日 | 1995-04-25 | | 公開日 | 1996-01-29 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Crystal structures of cyanide- and triiodide-bound forms of Arthromyces ramosus peroxidase at different pH values. Perturbations of active site residues and their implication in enzyme catalysis.
J.Biol.Chem., 270, 1995
|
|
1ARW
 
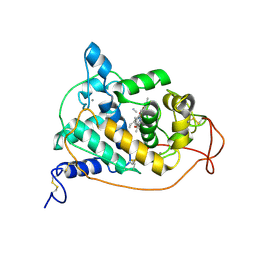 | | CRYSTAL STRUCTURES OF CYANIDE-AND TRIIODIDE-BOUND FORMS OF ARTHROMYCES RAMOSUS PEROXIDASE AT DIFFERENT PH VALUES. PERTURBATIONS OF ACTIVE SITE RESIDUES AND THEIR IMPLICATION IN ENZYME CATALYSIS | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CYANIDE ION, ... | | 著者 | Fukuyama, K, Kunishima, N, Amada, F. | | 登録日 | 1995-04-25 | | 公開日 | 1996-01-29 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Crystal structures of cyanide- and triiodide-bound forms of Arthromyces ramosus peroxidase at different pH values. Perturbations of active site residues and their implication in enzyme catalysis.
J.Biol.Chem., 270, 1995
|
|
1ARV
 
 | | CRYSTAL STRUCTURES OF CYANIDE-AND TRIIODIDE-BOUND FORMS OF ARTHROMYCES RAMOSUS PEROXIDASE AT DIFFERENT PH VALUES. PERTURBATIONS OF ACTIVE SITE RESIDUES AND THEIR IMPLICATION IN ENZYME CATALYSIS | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CYANIDE ION, ... | | 著者 | Fukuyama, K, Kunishima, N, Amada, F. | | 登録日 | 1995-04-25 | | 公開日 | 1996-01-29 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Crystal structures of cyanide- and triiodide-bound forms of Arthromyces ramosus peroxidase at different pH values. Perturbations of active site residues and their implication in enzyme catalysis.
J.Biol.Chem., 270, 1995
|
|
1ARX
 
 | | CRYSTAL STRUCTURES OF CYANIDE-AND TRIIODIDE-BOUND FORMS OF ARTHROMYCES RAMOSUS PEROXIDASE AT DIFFERENT PH VALUES. PERTURBATIONS OF ACTIVE SITE RESIDUES AND THEIR IMPLICATION IN ENZYME CATALYSIS | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, IODIDE ION, ... | | 著者 | Fukuyama, K, Kunishima, N, Amada, F. | | 登録日 | 1995-04-25 | | 公開日 | 1996-01-29 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structures of cyanide- and triiodide-bound forms of Arthromyces ramosus peroxidase at different pH values. Perturbations of active site residues and their implication in enzyme catalysis.
J.Biol.Chem., 270, 1995
|
|
