2LX4
 
 | |
3I4L
 
 | | Structural characterization for the nucleotide binding ability of subunit A with AMP-PNP of the A1AO ATP synthase | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, A-TYPE ATP SYNTHASE CATALYTIC SUBUNIT A, ... | | 著者 | Manimekalai, S.M.S, Kumar, A, Balakrishna, A.M, Jeyakanthan, J, Gruber, G. | | 登録日 | 2009-07-01 | | 公開日 | 2010-01-12 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Nucleotide binding states of subunit A of the A-ATP synthase and the implication of P-loop switch in evolution.
J.Mol.Biol., 396, 2010
|
|
3I73
 
 | | Structural characterization for the nucleotide binding ability of subunit A with ADP of the A1AO ATP synthase | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, A-TYPE ATP SYNTHASE CATALYTIC SUBUNIT A, ... | | 著者 | Manimekalai, S.M.S, Kumar, A, Balakrishna, A.M, Gruber, G. | | 登録日 | 2009-07-08 | | 公開日 | 2010-01-12 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Nucleotide binding states of subunit A of the A-ATP synthase and the implication of P-loop switch in evolution.
J.Mol.Biol., 396, 2010
|
|
4IX9
 
 | | Crystal structure of subunit F of V-ATPase from S. cerevisiae | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, V-type proton ATPase subunit F | | 著者 | Basak, S, Balakrishna, A.M, Manimekalai, M.S.S, Gruber, G. | | 登録日 | 2013-01-24 | | 公開日 | 2013-03-20 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.33 Å) | | 主引用文献 | Crystal and NMR structures give insights into the role and dynamics of subunit F of the eukaryotic V-ATPase from Saccharomyces cerevisiae
J.Biol.Chem., 288, 2013
|
|
3IKJ
 
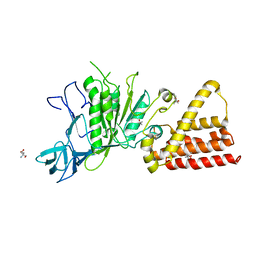 | | Structural characterization for the nucleotide binding ability of subunit A mutant S238A of the A1AO ATP synthase | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, V-type ATP synthase alpha chain | | 著者 | Kumar, A, Manimekali, M.S.S, Balakrishna, A.M, Jeyakanthan, J, Gruber, G. | | 登録日 | 2009-08-06 | | 公開日 | 2010-01-12 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Nucleotide binding states of subunit A of the A-ATP synthase and the implication of P-loop switch in evolution.
J.Mol.Biol., 396, 2010
|
|
3I72
 
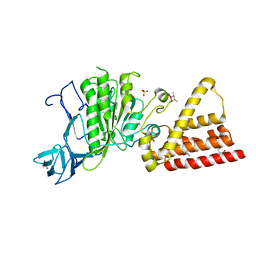 | | Structural characterization for the nucleotide binding ability of subunit A with SO4 of the A1AO ATP synthase | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, A-TYPE ATP SYNTHASE CATALYTIC SUBUNIT A, ... | | 著者 | Manimekalai, S.M.S, Kumar, A, Balakrishna, A.M, Gruber, G. | | 登録日 | 2009-07-07 | | 公開日 | 2010-01-12 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.47 Å) | | 主引用文献 | Nucleotide binding states of subunit A of the A-ATP synthase and the implication of P-loop switch in evolution.
J.Mol.Biol., 396, 2010
|
|
7DOJ
 
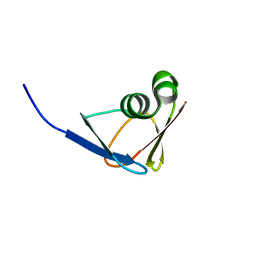 | |
8HGX
 
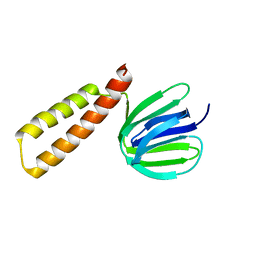 | |
3NI6
 
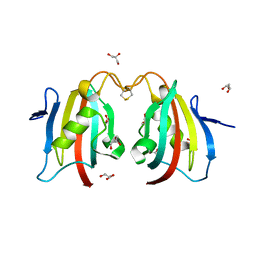 | |
5XGB
 
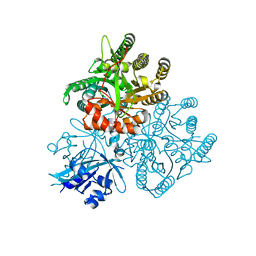 | |
5XGE
 
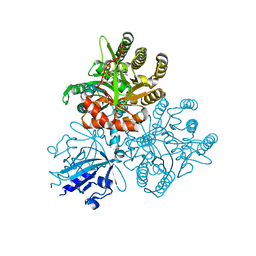 | | Crystal structure of the PAS-GGDEF-EAL domain of PA0861 from Pseudomonas aeruginosa in complex with cyclic di-GMP | | 分子名称: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Uncharacterized protein PA0861 | | 著者 | Liu, C, Liew, C.W, Sreekanth, R, Lescar, J. | | 登録日 | 2017-04-13 | | 公開日 | 2017-12-20 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (3.31 Å) | | 主引用文献 | Insights into Biofilm Dispersal Regulation from the Crystal Structure of the PAS-GGDEF-EAL Region of RbdA from Pseudomonas aeruginosa.
J. Bacteriol., 200, 2018
|
|
2OV6
 
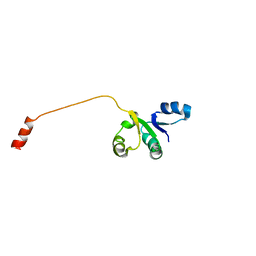 | |
5Y63
 
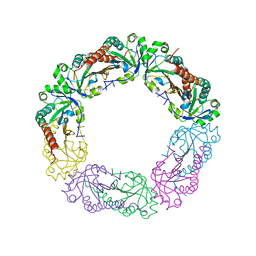 | |
7VIL
 
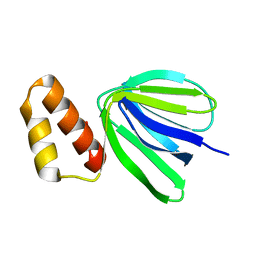 | |
7XKZ
 
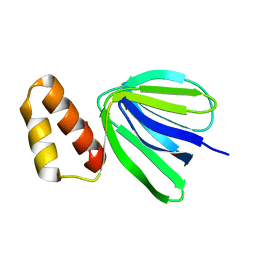 | | Solution structure of subunit epsilon of the Mycobacterium abscessus F-ATP synthase | | 分子名称: | ATP synthase epsilon chain | | 著者 | Shin, J, Grueber, G, Harikishore, A, Wong, C.F, Prya, R, Dick, T. | | 登録日 | 2022-04-20 | | 公開日 | 2023-03-08 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Atomic solution structure of Mycobacterium abscessus F-ATP synthase subunit epsilon and identification of Ep1MabF1 as a targeted inhibitor.
Febs J., 289, 2022
|
|
7Y5C
 
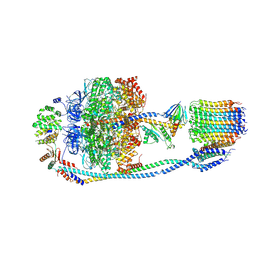 | | Cryo-EM structure of F-ATP synthase from Mycolicibacterium smegmatis (rotational state 2) | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | 著者 | Saw, W.-G, Wong, C.F, Grueber, G. | | 登録日 | 2022-06-16 | | 公開日 | 2022-11-23 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (4.7 Å) | | 主引用文献 | Structural Elements Involved in ATP Hydrolysis Inhibition and ATP Synthesis of Tuberculosis and Nontuberculous Mycobacterial F-ATP Synthase Decipher New Targets for Inhibitors.
Antimicrob.Agents Chemother., 66, 2022
|
|
7Y5A
 
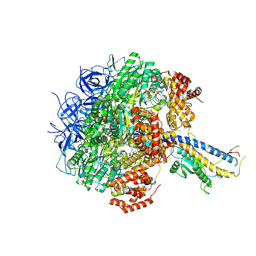 | | Cryo-EM structure of the Mycolicibacterium smegmatis F1-ATPase | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase gamma chain, ... | | 著者 | Wong, C.F, Saw, W.-G, Grueber, G. | | 登録日 | 2022-06-16 | | 公開日 | 2022-11-23 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Structural Elements Involved in ATP Hydrolysis Inhibition and ATP Synthesis of Tuberculosis and Nontuberculous Mycobacterial F-ATP Synthase Decipher New Targets for Inhibitors.
Antimicrob.Agents Chemother., 66, 2022
|
|
7Y5D
 
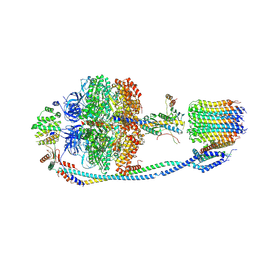 | | Cryo-EM structure of F-ATP synthase from Mycolicibacterium smegmatis (rotational state 3) (backbone) | | 分子名称: | ATP synthase epsilon chain, ATP synthase gamma chain, ATP synthase subunit a, ... | | 著者 | Saw, W.-G, Wong, C.F, Grueber, G. | | 登録日 | 2022-06-16 | | 公開日 | 2022-11-23 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (7.3 Å) | | 主引用文献 | Structural Elements Involved in ATP Hydrolysis Inhibition and ATP Synthesis of Tuberculosis and Nontuberculous Mycobacterial F-ATP Synthase Decipher New Targets for Inhibitors.
Antimicrob.Agents Chemother., 66, 2022
|
|
7Y5B
 
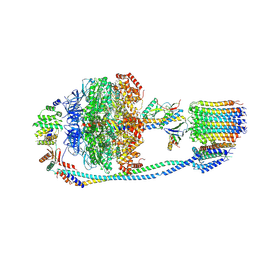 | | Cryo-EM structure of F-ATP synthase from Mycolicibacterium smegmatis (rotational state 1) | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | 著者 | Saw, W.-G, Wong, C.F, Grueber, G. | | 登録日 | 2022-06-16 | | 公開日 | 2022-11-23 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (4.4 Å) | | 主引用文献 | Structural Elements Involved in ATP Hydrolysis Inhibition and ATP Synthesis of Tuberculosis and Nontuberculous Mycobacterial F-ATP Synthase Decipher New Targets for Inhibitors.
Antimicrob.Agents Chemother., 66, 2022
|
|
7CK1
 
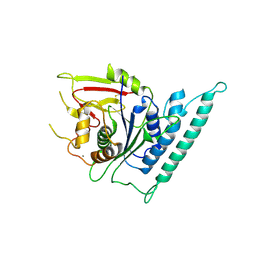 | | Crystal structure of arabidopsis CESA3 catalytic domain | | 分子名称: | Cellulose synthase A catalytic subunit 3 [UDP-forming],Cellulose synthase A catalytic subunit 3 [UDP-forming], MANGANESE (II) ION | | 著者 | Qiao, Z, Gao, Y.G. | | 登録日 | 2020-07-15 | | 公開日 | 2021-03-17 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Structure of Arabidopsis CESA3 catalytic domain with its substrate UDP-glucose provides insight into the mechanism of cellulose synthesis.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7CK3
 
 | | Crystal structure of Arabidopsis CESA3 catalytic domain | | 分子名称: | Cellulose synthase A catalytic subunit 3 [UDP-forming],Cellulose synthase A catalytic subunit 3 [UDP-forming] | | 著者 | Qiao, Z, Gao, Y.G. | | 登録日 | 2020-07-15 | | 公開日 | 2021-03-17 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structure of Arabidopsis CESA3 catalytic domain with its substrate UDP-glucose provides insight into the mechanism of cellulose synthesis.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7CK2
 
 | | Crystal structure of Arabidopsis CESA3 catalytic domain with UDP-Glucose | | 分子名称: | Cellulose synthase A catalytic subunit 3 [UDP-forming],Cellulose synthase A catalytic subunit 3 [UDP-forming], MANGANESE (II) ION, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | 著者 | Qiao, Z, Gao, Y.G. | | 登録日 | 2020-07-15 | | 公開日 | 2021-03-17 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Structure of Arabidopsis CESA3 catalytic domain with its substrate UDP-glucose provides insight into the mechanism of cellulose synthesis.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
4PPI
 
 | | Crystal structure of Bcl-xL hexamer | | 分子名称: | Bcl-2-like protein 1, GLYCEROL | | 著者 | Sreekanth, R, Yoon, H.S. | | 登録日 | 2014-02-27 | | 公開日 | 2015-03-04 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.851 Å) | | 主引用文献 | Structural transition in Bcl-xL and its potential association with mitochondrial calcium ion transport
Sci Rep, 5, 2015
|
|
5XGD
 
 | | Crystal structure of the PAS-GGDEF-EAL domain of PA0861 from Pseudomonas aeruginosa in complex with GTP | | 分子名称: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Uncharacterized protein PA0861 | | 著者 | Liu, C, Liew, C.W, Sreekanth, R, Lescar, J. | | 登録日 | 2017-04-13 | | 公開日 | 2017-12-20 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Insights into Biofilm Dispersal Regulation from the Crystal Structure of the PAS-GGDEF-EAL Region of RbdA from Pseudomonas aeruginosa.
J. Bacteriol., 200, 2018
|
|
5W41
 
 | |
