7XS8
 
 | |
7XSA
 
 | |
7XSB
 
 | |
7XSC
 
 | |
7XN9
 
 | | Crystal structure of SSTR2 and L-054,522 complex | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Somatostatin receptor type 2,Endo-1,4-beta-xylanase, tert-butyl (2S)-6-azanyl-2-[[(2R,3S)-3-(1H-indol-3-yl)-2-[[4-(2-oxidanylidene-3H-benzimidazol-1-yl)piperidin-1-yl]carbonylamino]butanoyl]amino]hexanoate | | 著者 | Zhao, W, Han, S, Qiu, N, Feng, W, Lu, M, Yang, D, Wang, M.-W, Wu, B, Zhao, Q. | | 登録日 | 2022-04-28 | | 公開日 | 2022-08-03 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structural insights into ligand recognition and selectivity of somatostatin receptors.
Cell Res., 32, 2022
|
|
7XNA
 
 | | Crystal structure of somatostatin receptor 2 (SSTR2) with peptide antagonist CYN 154806 | | 分子名称: | CYN 154806, Somatostatin receptor type 2,Endo-1,4-beta-xylanase | | 著者 | Zhao, W, Han, S, Qiu, N, Feng, W, Lu, M, Yang, D, Wang, M.-W, Wu, B, Zhao, Q. | | 登録日 | 2022-04-28 | | 公開日 | 2022-08-03 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Structural insights into ligand recognition and selectivity of somatostatin receptors.
Cell Res., 32, 2022
|
|
5ZO0
 
 | |
7V4X
 
 | |
7V51
 
 | | BVMO_negative mutant D432V | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, Putative flavin-binding monooxygenase | | 著者 | Wu, Y, Yu, H.-L. | | 登録日 | 2021-08-16 | | 公開日 | 2023-02-22 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.42 Å) | | 主引用文献 | Precise regulation of the substrate selectivity of Baeyer-Villiger monooxygenase to minimize overoxidation of prazole sulfoxides.
CHINESE J CATAL, 51, 2023
|
|
7V50
 
 | |
7EMN
 
 | |
7FC3
 
 | | structure of NL63 receptor-binding domain complexed with horse ACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ... | | 著者 | Wang, X.Q, Ge, J.W, Lan, J. | | 登録日 | 2021-07-13 | | 公開日 | 2021-09-22 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.19 Å) | | 主引用文献 | Structural insights into the binding of SARS-CoV-2, SARS-CoV, and hCoV-NL63 spike receptor-binding domain to horse ACE2.
Structure, 30, 2022
|
|
7FC5
 
 | | Crystal structure of SARS-CoV-2 RBD and horse ACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, Spike protein S1 | | 著者 | Wang, X.Q, Lan, J, Ge, J.W. | | 登録日 | 2021-07-13 | | 公開日 | 2022-06-22 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.894 Å) | | 主引用文献 | Structural insights into the binding of SARS-CoV-2, SARS-CoV, and hCoV-NL63 spike receptor-binding domain to horse ACE2.
Structure, 30, 2022
|
|
7FC6
 
 | | Crystal structure of SARS-CoV RBD and horse ACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ... | | 著者 | Wang, X.Q, Lan, J, Ge, J.W. | | 登録日 | 2021-07-13 | | 公開日 | 2022-07-13 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.655 Å) | | 主引用文献 | Structural insights into the binding of SARS-CoV-2, SARS-CoV, and hCoV-NL63 spike receptor-binding domain to horse ACE2.
Structure, 30, 2022
|
|
7F29
 
 | |
8C8H
 
 | |
2JVA
 
 | | NMR solution structure of peptidyl-tRNA hydrolase domain protein from Pseudomonas syringae pv. tomato. Northeast Structural Genomics Consortium target PsR211 | | 分子名称: | Peptidyl-tRNA hydrolase domain protein | | 著者 | Singarapu, K.K, Sukumaran, D, Parish, D, Eletsky, A, Zhang, Q, Zhao, L, Jiang, M, Maglaqui, M, Xiao, R, Liu, J, Baran, M.C, Swapna, G.V.T, Huang, Y.J, Acton, T.B, Rost, B, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2007-09-14 | | 公開日 | 2007-10-02 | | 最終更新日 | 2024-05-08 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR structure of the peptidyl-tRNA hydrolase domain from Pseudomonas syringae expands the structural coverage of the hydrolysis domains of class 1 peptide chain release factors.
Proteins, 71, 2008
|
|
7FAE
 
 | | S protein of SARS-CoV-2 in complex bound with P36-5D2(state2) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, P36-5D2 heavy chain, ... | | 著者 | Zhang, L, Wang, X, Shan, S, Zhang, S. | | 登録日 | 2021-07-06 | | 公開日 | 2021-12-22 | | 最終更新日 | 2022-02-16 | | 実験手法 | ELECTRON MICROSCOPY (3.65 Å) | | 主引用文献 | A Potent and Protective Human Neutralizing Antibody Against SARS-CoV-2 Variants.
Front Immunol, 12, 2021
|
|
7FAF
 
 | | S protein of SARS-CoV-2 in complex bound with P36-5D2 (state1) | | 分子名称: | P36-5D2 heavy chain, P36-5D2 light chain, Spike glycoprotein | | 著者 | Zhang, L, Wang, X, Zhang, S, Shan, S. | | 登録日 | 2021-07-06 | | 公開日 | 2021-12-22 | | 最終更新日 | 2022-02-16 | | 実験手法 | ELECTRON MICROSCOPY (3.69 Å) | | 主引用文献 | A Potent and Protective Human Neutralizing Antibody Against SARS-CoV-2 Variants.
Front Immunol, 12, 2021
|
|
7WTK
 
 | | SARS-CoV-2 Omicron variant spike in complex with Fab XGv286 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of XGv286, ... | | 著者 | Wang, X, Fu, W. | | 登録日 | 2022-02-04 | | 公開日 | 2022-12-14 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Selection and structural bases of potent broadly neutralizing antibodies from 3-dose vaccinees that are highly effective against diverse SARS-CoV-2 variants, including Omicron sublineages.
Cell Res., 32, 2022
|
|
7WTJ
 
 | | SARS-CoV-2 Omicron variant spike RBD in complex with Fab XGv286 | | 分子名称: | Heavy chain of XGv286, Light chain of XGv286, Spike protein S1 | | 著者 | Wang, X, Fu, W. | | 登録日 | 2022-02-04 | | 公開日 | 2022-12-14 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | Selection and structural bases of potent broadly neutralizing antibodies from 3-dose vaccinees that are highly effective against diverse SARS-CoV-2 variants, including Omicron sublineages.
Cell Res., 32, 2022
|
|
7WTI
 
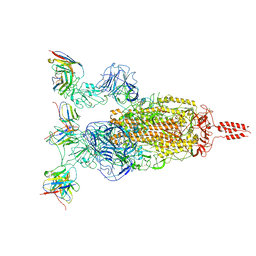 | | SARS-CoV-2 Omicron variant spike in complex with Fab XGv264 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of XGv264, Light chain of XGv264, ... | | 著者 | Wang, X, Fu, W. | | 登録日 | 2022-02-04 | | 公開日 | 2022-12-21 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Selection and structural bases of potent broadly neutralizing antibodies from 3-dose vaccinees that are highly effective against diverse SARS-CoV-2 variants, including Omicron sublineages.
Cell Res., 32, 2022
|
|
7WTF
 
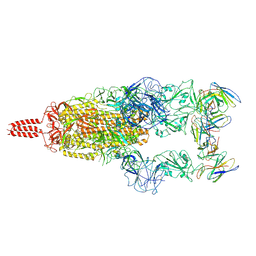 | | SARS-CoV-2 Omicron variant spike in complex with Fab XGv051 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of XGv051, ... | | 著者 | Wang, X, Fu, W. | | 登録日 | 2022-02-04 | | 公開日 | 2022-12-14 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Selection and structural bases of potent broadly neutralizing antibodies from 3-dose vaccinees that are highly effective against diverse SARS-CoV-2 variants, including Omicron sublineages.
Cell Res., 32, 2022
|
|
7WTG
 
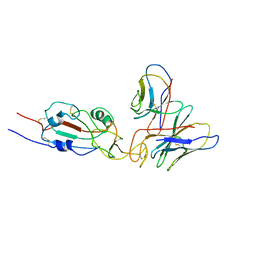 | | SARS-CoV-2 Omicron variant spike RBD in complex with Fab XGv051 | | 分子名称: | Heavy chain of XGv051, Light chain of XGv051, Spike protein S1 | | 著者 | Wang, X, Fu, W. | | 登録日 | 2022-02-04 | | 公開日 | 2022-12-14 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Selection and structural bases of potent broadly neutralizing antibodies from 3-dose vaccinees that are highly effective against diverse SARS-CoV-2 variants, including Omicron sublineages.
Cell Res., 32, 2022
|
|
7WTH
 
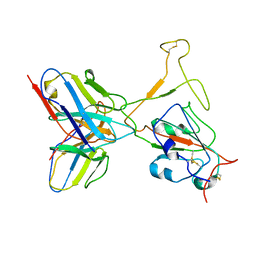 | | SARS-CoV-2 Omicron variant spike RBD in complex with Fab XGv264 | | 分子名称: | Heavy chain of XGv264, Light chain of XGv264, Spike protein S1 | | 著者 | Wang, X, Fu, W. | | 登録日 | 2022-02-04 | | 公開日 | 2022-12-14 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | Selection and structural bases of potent broadly neutralizing antibodies from 3-dose vaccinees that are highly effective against diverse SARS-CoV-2 variants, including Omicron sublineages.
Cell Res., 32, 2022
|
|
